- Title
-
The RNA Binding Protein Igf2bp1 Is Required for Zebrafish RGC Axon Outgrowth In Vivo
- Authors
- Gaynes, J.A., Otsuna, H., Campbell, D.S., Manfredi, J.P., Levine, E.M., Chien, C.B.
- Source
- Full text @ PLoS One
|
The β-actin 3′UTR is sufficient for local translation of Kaede in RGC growth cones in vivo. (a) Steps for in vivo timelapse assay: 1- cDNA injections, 2- selection of embryos with strong Kaede expression in the eye at 2dpf, 3- dissections to remove right eye and drain yolk, 4- photoconversion of Kaede in RGC axons, 5- timelapse. (b) Confocal projection (40x water lens) of a live 3 dpf embryo with Kaede expression in RGC axons in the optic tract (green). (c-f) Confocal projections of the green and red channels of one axon before (c, d) and after photoconversion (e, f). (g-n) Confocal projection from timelapse of one–UTR axon (g-j) and one +UTR axon (k-n) with green to red fluorescence ratio represented by color map. (o, p) Average green to red fluorescence intensity ratio throughout timelapse in growth cones (pixels 1–10) and proximal regions (pixels 141–150) of–UTR (n = 6) and +UTR (n = 10) axons. A one-way ANOVA (p = 0.0012) with a Tukey HSD test (p<0.05) 90 min after photoconversion showed significantly higher green to red ratio in +UTR growth cones compared to–UTR growth cones at 90 min after photoconversion. (q) The green-to-red ratio in a representative +UTR axon (axon 1) plotted against the distance from the growth cone at a representative time (90 min) after photoconversion. The slope of a linear regression to these data (outlined with a magenta rectangle) reflects the spatial gradient of green-to-red ratio 90 minutes after photoconversion. (r) Change in spatial gradient of green-to-red ratio in representative axon 1 throughout the timelapse. The point labeled Q is the slope of the linear regression in panel q, which was determined at 90 min. The other points in this graph were similarly determined in axon 1 at 10 minute intervals throughout the timelapse assay. The slope of a linear regression to these data (outlined with a green rectangle) reflects the rate of change of the gradient of green-to-red fluorescence along axon 1. (s) Rates of change in gradients of green-to-red fluorescence in all assayed axons. The point labeled R is the slope of the linear regression in panel r, which was determined in axon 1. The rates of change of the axonal gradient for the UTR (n = 6) and +UTR (n = 10) axons were significantly different (Mann-Whitney U test, p = 0.0002). Scale bars are 100 µm (b) and 5 µm (g). |
|
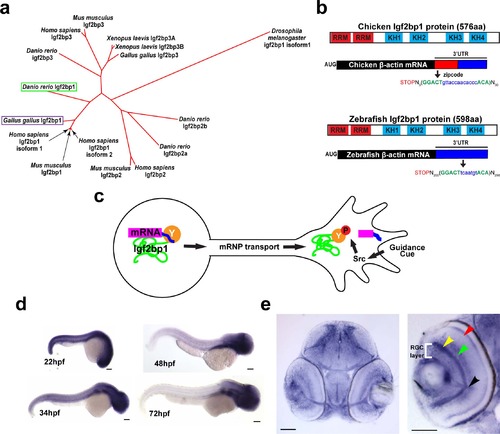
Igf2bp1, the zebrafish ZBP1 ortholog, is expressed in RGCs during axon pathfinding. (a) Phylogenetic tree generated with ClustalW, showing that of the four Igf2bp genes in Danio rerio Igf2bp1 is the most closely related to Gallus gallus Igf2bp1 of the four Igf2bp genes in Danio rerio, based on amino acid sequence. b) Schematic of zebrafish and chicken Igf2bp1 and β-actin mRNA, illustrating the conservation of protein domains and the presence of a bipartite nucleotide sequence in Danio rerio mRNA that is homologous to the zipcode sequence of chicken. These similarities at both the protein and mRNA levels suggest that in zebrafish, as has been shown in chicken, KH34 binds the putative zipcode sequence in the β-actin 3′UTR. (c) Schematic of mechanism for localization of β-actin mRNA by Igf2bp1 predicted to be conserved in zebrafish RGCs in vivo. (d) Wholemount in situ hybridizations performed on 22 hpf, 48 hpf, 34 hpf, and 72 hpf embryos show the location of Igf2bp1 mRNA expression. (e) 15 µm coronal plastic sections show Igf2bp1 mRNA expression in the RGC layer (white bracket in right panel), as well as the inner plexiform layer (yellow arrowhead), inner nuclear layer (green arrowhead), photoreceptor layer (red arrowhead) and RGC axons in the optic nerve (black arrowhead) in 3 dpf embryos. Scale bars are 100 µm (d) and 50 µm (e). EXPRESSION / LABELING:
|
|
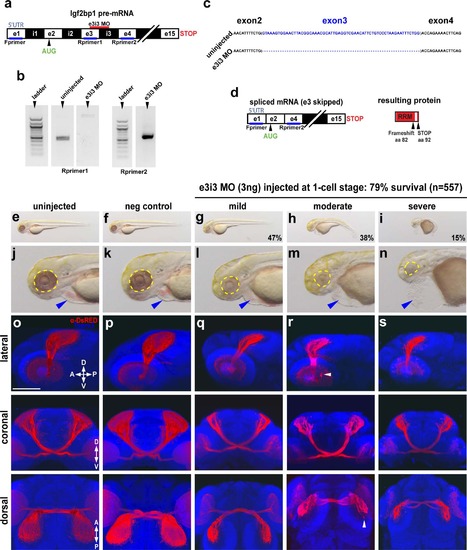
Impairment of Igf2bp1 perturbs retinotectal projections. (a) The e3i3 MO targeted to the e3i3 splice junction in Igf2bp1 pre-mRNA. (b) Left gel: forward primer targeted to exon 1 in the 5′UTR (Fprimer) and a reverse primer targeted to exon 3 (Rprimer1) yielded no detectable product as expected with exon 3 deletion. Right gel: Fprimer and a reverse primer targeted to exon 4 (Rprimer2) yielded a PCR product that was TOPO-TA cloned and sequenced (c) to verify exon 3 deletion (blank lanes were cropped out of gel image), which results in a severely truncated protein (d). (e-n) Transmitted light images of whole 3 dpf control (e,f) and e3i3 MO-injected morphants (g-i) and head regions (j-n) with eyes (yellow outline) and hearts (blue arrowheads). (o-s) 3D projections made from confocal z-stacks taken with a 30x silicone immersion lens on a confocal microscope, of Tg(isl2b:mCherryCAAX)zc23 3 dpf embryos stained with α-DsRed (red) and counterstained with TO-PRO-3 (blue), with one example each for uninjected (o), or injected with negative control MO (p), e3i3 MO (mild (q), moderate (r), severe (s)), reconstructed from confocal z-stacks with FluoRender software, rotated to give lateral (top panels), coronal (middle panels) and dorsal (bottom panels) views. Scale bar is 100 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Impairment of Igf2bp1 increases cell death and layering defects in the retina. Tg(isl2b:GFP)zc7 stable transgenic embryos (a-h) were uninjected (a, e) injected with p53 MO (b, f), e3i3 (c, g), or co-injected with p53 MO+e3i3 MO (d, h). Embryos were fixed and stained with α-EGFP (green) and TO-PRO-3 (red in a-d, gray in e-h). Images are maximum intensity projections (a-d, i-l) or singles slices (e-h) of lateral views of eyes, with lens removed, taken with a confocal microscope (20x lens). Morphant eyes were missing RGCs in central retina (white arrowheads in c, d) and had displaced RGCs (yellow arrowheads in c, d). (g, h) Single z-slices have holes (white arrowheads) and abnormal layers (yellow arrowheads). (i, l) Maximum intensity projections, lateral view of AO staining in 31 hpf embryos, within the lens (inner dotted yellow circle) and retina (outer dotted yellow circle). (m) The AO-positive cells were counted in retinas alone (not including lens and extraocular tissues) from; uninjected (n = 10), p53 MO injected (n = 9), e3i3 MO injected (n = 10), and coinjected with p53 MO and e3i3 MO (n = 10) embryos. A one-way ANOVA (p<0.0001) with Tukey HSD test (p<0.01) showed that e3i3 MO injected embryos and embryos coinjected with e3i3 MO and p53 MO were both significantly different than controls, but not significantly different from each other. The black points on the graph represent mean +/- SEM. Scale bars are 50 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression of emGFP-lgf2bp1Y399E in RGCs disrupts axon outgrowth in vivo. (a-c) Dorsal confocal projections (30x silicone immersion lens) of Tg(isl2b:emGFP) (a), Tg(isl2b:emGFP-Igf2bp1)wt (b), and Tg(isl2b:emGFP-Igf2bp1)Y399E (c) 3 dpf transient transgenic embryos. (d) Quantification of the ratio of emGFP (green) positive RGCs in the retina to emGFP positive axons on the contralateral tectum per tectum-eye pair (average +/- SEM). A one-way ANOVA (p<0.0001) with a Tukey HSD test (p<0.01) showed that the ratios were significantly different in emGFP-Igf2bp1Y399E (N = 16 tectum-eye pairs, 9 animals) compared to Igf2bp1wt (N = 11 tectum-eye pairs in 7 animals) and emGFP (N = 26 tectum-eye pairs, 16 animals) controls, and controls were not significantly different. (e) Schematic of the predicted mechanism for Igf2bp1Y399E expression preventing axon outgrowth by interfering with endogenous Igf2bp1 by competing for mRNA transport to the growth cone without β-actin mRNA cargo, therefore decreasing the amount of β-actin mRNA available for local translation in the growth cone in response to attractive guidance cues. (f-k) Dorsal confocal projections of Tg(isl2b:emGFP (f-h) and Tg(isl2b:emGFP-Igf2bp1Y399F) (i-k) 3 dpf embryos. Scale bars are 100 µm. |
|
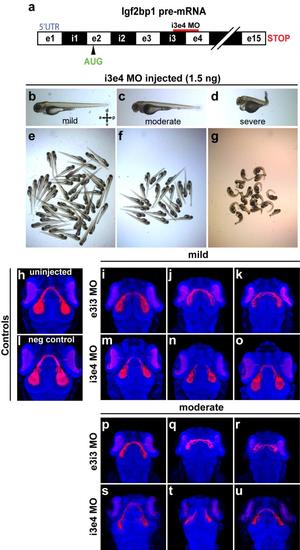
Injection with i3e4 MO gives retinotectal phenotype similar to e3i3 MO. (a) The i3e4 MO targeted to the i3e4 splice junction in Igf2bp1 pre-mRNA. (b-g) Transmitted light images of whole 3 dpf i3e4 MO-injected morphants. (h-u) 3D projections made from confocal z-stacks take with a 20x lens on a confocal microscope, of Tg(isl2b:mCherryCAAX)zc23 3 dpf embryos stained with α-DsRed (red) and counterstained with TO-PRO-3 (blue), with one example each for uninjected (h), or injected with negative control MO (l), and three examples each injected with e3i3 MO (mild (i-k), moderate (p-r)) or i3e4 MO (mild (m-o), moderate (s-u)). |
|
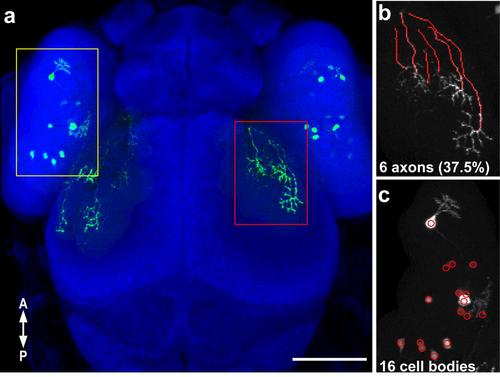
Quantification of the ratio of labeled RGC axons to labeled cell bodies in Fig 5. (a) Confocal projection (30x silicone immersion lens) of a 3 dpf embryo with transient expression of isl2b:emGFP, with labeled RGC axons (a red rectangle, b) and labeled cell bodies (yellow rectangle, c) that were counted. Axons were traced in the dorsal optic tract and tectum using the Fiji simple neurite tracer plugin (b). (c) The cell counter plugin in ImageJ was used to mark and count labeled cells in the contralateral retina. The ratio of labeled axons to labeled cell bodies per retinotectal projection was calculated (37.5%). Scale bar is 100 µm. |

Unillustrated author statements EXPRESSION / LABELING:
|