- Title
-
PIAS-like protein Zimp7 is required for the restriction of the Zebrafish organizer and mesoderm development
- Authors
- Moreno-Ayala, R., Schnabel, D., Salas-Vidal, E., Lomelí, H.
- Source
- Full text @ Dev. Biol.
|
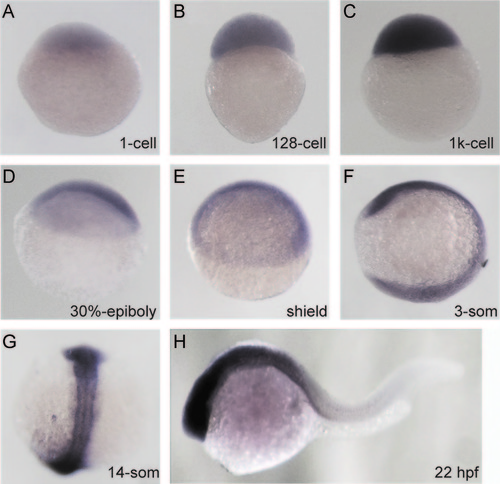
Spatiotemporal Expression Pattern of zimp7 Expression pattern of zimp7 detected by whole-mount in situ hybridization, at indicated stages. (A–E), lateral views with the animal pole oriented at the top; (F), lateral with anterior at the top; (G), dorsal view with anterior at the top (H), anterior is oriented towards the left. EXPRESSION / LABELING:
|
|
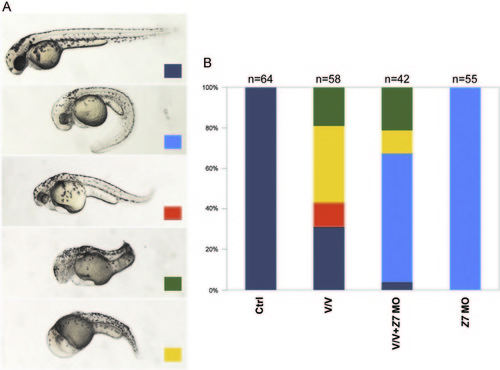
Zimp7 loss-of-function causes axial phenotypes. (A) Embryos injected with a control MO at 9 hpf; relative to this embryos. (B) zimp7-sp MO injected embryos present an elongated shape. (C) After 30 hpf of development, zimp7-sp MO injected embryos are ventrally curved. (D) Co-injection of embryos with the zimp7-sp MO and the p53 MO rescued head defects but not axial defects. (E) At 30 hpf embryos injected with a zimp7-ATG MO present a similar phenotype. (F) Embryos with specific mutations in the zimp7 gene produced by the CRISPR/Cas system present a curly down tail and a similar appearance as the zimp7-sp MO injected embryos. (G) Distribution of phenotypes produced with the injected morpholinos. Total numbers of embryos are shown at the top. PHENOTYPE:
|
|
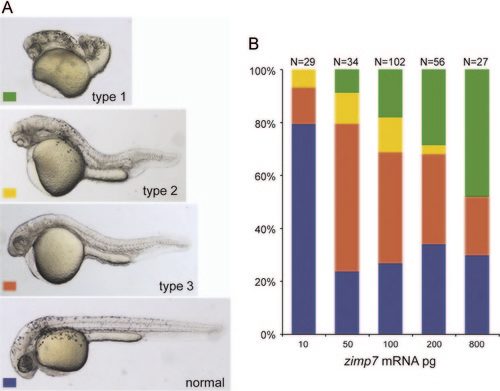
Zimp7 overexpression produces axial mesoderm defects. Injection of 10–800 pg of zimp7 mRNA produced malformations that were classified into four categories. (A) Type1 embryos are very severely defective. Type 2 embryos showed cyclopia, anterior truncations, bent tail and shortened body. Type 3 embryos presented a shortened body axis. Lateral views. (B) Distribution of embryonic malformations. Total numbers of embryos are shown at the top. Defects induced by zimp7 mRNA are dose dependent. |
|
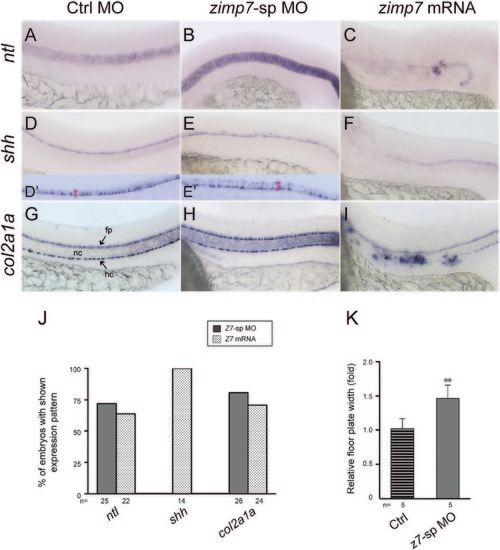
Loss and gain of function of Zimp7 exert opposite effects in midline structures. Lateral views (anterior is left) of ntl (A–C), shh (D–F) and col2a1a (G–I) expression in Ctrl MO+p53 MO (A, D, G), zimp7-sp MO+p53 MO (B, E, H) and zimp7 mRNA (C, F, I) injected embryos at 26 hpf. Compared to the control embryo (A), ntl expression in the notochord is increased in a Zimp7 knockdown embryo (B), while it is nearly lost in the gain-of-function embryo (C). Expression of shh marks the floor plate. Compared to the control embryo (D) the floor plate is widened in the morphant (E). D′ and E′ show four-times magnified images of the corresponding embryos, where the engrossment of the floor plate was measured (area indicated with a red bar). shh expression reveals the lost of floor plate in embryos injected with a zimp7 mRNA (F). Expression of col2a1a outlines the floor plate and hypochord in a control embryo (G). (H) In knockdown embryos, no significant differences are appreciated in the hypochord, whilst the floor plate exhibits a stronger signal compared to the control embryo. In the embryos overexpressing Zimp7 (I) the signal in the hypochord is defective and ectopic positive signal is detected. fp, floor plate; hc, hypochord; nc, notochord. (J) Percentage of embryos with shown expression pattern in B, C, F and I. Total number of embryos examined indicated at the bottom. (K) Measurements of shh positive signal in Ctrl MO and zimp7-sp MO injected embryos. **p<0.01. |
|
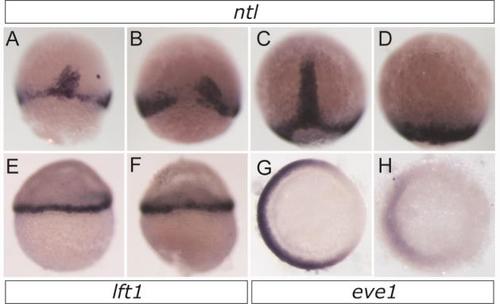
Zimp7 restricts organizer gene expression. Dorsal and ventral gene expression in embryos at shield (A–N), 5 hpf (O–Q) and 3-somite stage (R–T). Embryos were hybridized to specific marker genes boz (A and B), gsc (C–E), flh (F–H), vox (I–K), vent (L–N), ntl (O–Q) and hgg1 (R–T). The injected reagents are indicated at the left. boz, lateral views, dorsal to the right; gsc, and flh dorsal views with the animal pole oriented toward the top; vox, vent and ntl, animal pole views with dorsal to the right; hgg1, anterior view, dorsal side to the top. Arrows in I–N indicate the angle of marginal expression of ventral genes. An asterisk indicates the ectopic expression of vent in the ectoderm. U and V, graphs showing average values obtained from a number of embryos (indicated at the bottom of each bar) as the ones shown in A–T. U, the relative differences in mean expression area in MO and mRNA injected embryos compared to control embryos (horizontal line normalized to one) for the dorsal genes and hgg1. V, the average negative expression angle in MO and mRNA injected embryos compared to control embryos for the ventral genes vox and vent. The number of embryos with given appearance for ntl (Q) is 81% (n=21). zimp7 morphants display a significant increase in all dorsal genes as well as in the organizer-induced gene hgg1 and a reduction in ventral gene expression. Opposite effects are observed when zimp7 is overexpressed. Notably, besides an increased marginal angle in ventral genes, there is augmented ectodermal expression. Np<0.05; **p<0.01; ***p<0.001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
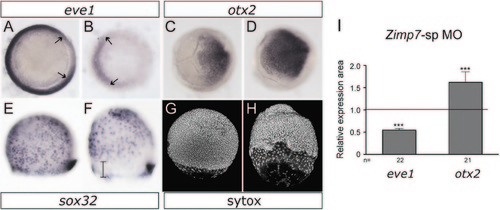
Analysis of markers at mid-gastrulation. 75% Epiboly stage embryos. (A and B) Decreased Zimp7 activity causes a reduced signal in the ventrolateral mesoderm marker eve1 (arrows), and up-regulation of the neuroectodermal marker otx2 (C and D). (E and F) Expression of the endoderm and dorsal forerunner cells (DFCs) marker, sox32. Although comparable numbers of endodermal cells are detected in the morphants, the endodermal precursor cells are delayed relative to the DFCs (bar), which remain normal. (G and H) Sytox stained embryos. Similar migration defects are observed, and a bigger gap between the deep layer cells and the enveloping layer cells are observed in the zimp7 morphants relative to the control. eve1, vegetal views, dorsal to the right. otx2 animal views, dorsal to the right. (E–H) lateral views, animal to the top. (A, C, E, and G) control embryos and (B, D, F, and H) zimp7 morphants are shown. I, total number of embryos examined (indicated at the bottom) and the mean expression area differences between Ctrl MO and zimp7-sp MO injected embryos shown in A–D. The number of embryos with given appearance for sox32 (F) is 77% (n=22) and for Sytox 100% (n=5). ***p<0.001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Co-injection of zimp7-sp MO and mRNA restores the normal level of expression of markers in embryos. (A) Anterior views of 3-somite stage embryos with dorsal toward the top. Expression of hgg1 is shown for control embryos, zimp7-sp MO injected embryos and embryos co-injected with zimp7-sp MO plus the indicated amounts of zimp7 mRNA. (B) Expression of gsc at shield stage in control embryos, zimp7-sp MO injected embryos and embryos co-injected with zimp7-sp MO plus the indicated amounts of zimp7 mRNA. Dorsal views. (C and D) For both markers the expression area was measured for a group of embryos (numbers indicated at the bottom of each bar) and the average area was plotted for each condition. Statistical analysis indicated a significant rescue of the normal level of expression for both markers. Bars represent standard deviation. **p<0.01; ***p<0.001. EXPRESSION / LABELING:
|
|
Reduction of Zimp7 activity suppresses the loss of head detected in Vox and Vent mediated ventralizations. (A) Dark-blue signed embryos are normal looking ones. Injection of 8 ng of the zimp7-sp MO+p53 MO produces embryos with the axial-dorsal phenotype indicated with a light-blue sign. Co-injection of vox and vent mRNAs produces morphologies displayed and classified in the pictures with orange, green and yellow signs. Orange indicates embryos with mild head defects and a bent tail. Green signed embryos present forebrain loss and shortened axis; yellow signed embryos present a V3-like morphology including the loss of head. (B) A graph showing fractions of embryos at 52 hpf that displayed the indicated phenotypes. PHENOTYPE:
|
|
Embryos with a severe phenotype produced by the zimp7-sp MO injection (A), the zimp7-sp MO and the p53 MO co-injection (B) and the Cas9/zimp7-gRNA injection (C). (D) PCR amplification of zimp7 exons 1 to 4 confirm the splicing alterations produced by the zimp7-sp MO. Total RNA was purified from embryos injected with control and zimp7-sp MO. The 522 bp fragment corresponds to the amplicon produced by the regular transcript (including exons 1, 2, 3 and 4); the 407 bp fragment is produced when exon 2 is not included in the transcript. An additional unexpected 485 bp fragment was obtained. Sequencing of this fragment revealed a cryptic splice site within exon 2, which generated a fragment that partially lost this exon. Both shorter species generate out-of-frame products and non-functional peptides. Densitometric analysis indicated that the shorter fragments combined represent 60% of the total RNA. (E) A T7-endonuclease assay. PCR product samples from pools of Cas9/zimp7-gRNA injected embryos with zimp7-like phenotypes (Phen) or without defects (Normal) were digested for 20 minutes with the T7 EI. Comparison of the digested products revealed the presence of more mutations in the zimp7-like embryos. The control sample is a PCR product from non-injected embryos digested with T7. (F) DNA sequence of samples obtained from embryos with abnormalities demonstrates the presence of specific mutations in the zimp7 gene. The target sequence is highlighted (PAM in green). PHENOTYPE:
|
|
Embryos overexpressing zimp7 presented eye defects including synophtalmia (B), one-eye loss (C) and cyclopia (D). |
|
Expression of ntl in 26 hpf embryos (A-C) Whole bodies of embryos presented in Fig. 4. In these images the extension of the expression of ntl along the notochord can be better appreciated. Embryos were injected with the indicated reagent. EXPRESSION / LABELING:
|
|
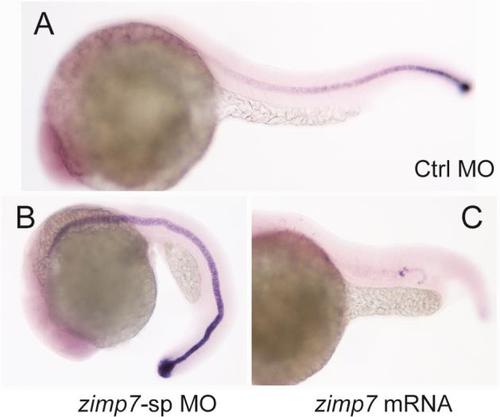
Alterations on Zimp7 levels produce abnormal expression of ntl and eve1, but not of lft1. Expression of ntl was detected by WISH in wild type (A,C) or embryos injected with zimp7 mRNA (B,D). (A-B) 65% (n = 73) of embryos at shield-stage and 56% (n = 30) of embryos at 90%-epiboly (C-D) lost dorsal expression upon zimp7 mRNA injection. Dorsal views, animal pole toward the top. Embryos injected with Ctrl MO (E,G) and zimp7-sp MO (F,H). (E-F) lft1, lateral views, animal pole toward the top, shield stage. (G-H) Expression of eve1 at 75%-epiboly, vegetal views with dorsal side at the right (a 56% decrease in the expression area was observed, n=19). EXPRESSION / LABELING:
|
|
Dorsal expression of boz in Ctrl MO (A) and zimp7-sp MO (B) 4 hpf. No obvious differences were appreciated. (B) Relative quantification of transcripts calculated from triplicate PCR samples. Changes of expression relative to control embryos (fold change =1) of sqt, boz, gsc, vox and vent in zimp7-MO injected embryos at 4 hpf. Statistical analysis indicated that changes were not significant EXPRESSION / LABELING:
|
|
Supplementary materialSupplementary Figure 6 (A-B) Changes of Zimp7 activity do not compensate variations in Nodal activity. (A) Injection of 3 ng of the oep MO, which decreases the expression of the Nodal-signaling co-receptor Oep, produces characteristic Nodal loss-of-function phenotypes (classified according to Kapp et al). The reduction of Zimp7 activity by co-injecting the oep MO with 4 ng of the zimp7-sp MO does not significantly affect the distribution of phenotypes. (B) Injection of 2pg of the sqt mRNA produces dorsalizations that are not alleviated by the co-injection of 80 or 120 pg of the zimp7 mRNA. (C) Injection of 2 ng of a chd MO previously used to increase BMP activity produces ventralizations, which are not rescued by a reduction of Zimp7 activity with 8 ng of the zimp7-sp MO. Morpholinos were coinjected with p53 MO to score the embryos for phenotype at 30hpf. PHENOTYPE:
|
Reprinted from Developmental Biology, 403(1), Moreno-Ayala, R., Schnabel, D., Salas-Vidal, E., Lomelí, H., PIAS-like protein Zimp7 is required for the restriction of the Zebrafish organizer and mesoderm development, 89-100, Copyright (2015) with permission from Elsevier. Full text @ Dev. Biol.