- Title
-
Motor neuron-expressed microRNAs 218 and their enhancers are nested within introns of SLIT2/3 genes
- Authors
- Punnamoottil, B., Rinkwitz, S., Giacomotto, J., Svahn, A.J., Becker, T.S.
- Source
- Full text @ Genesis
|
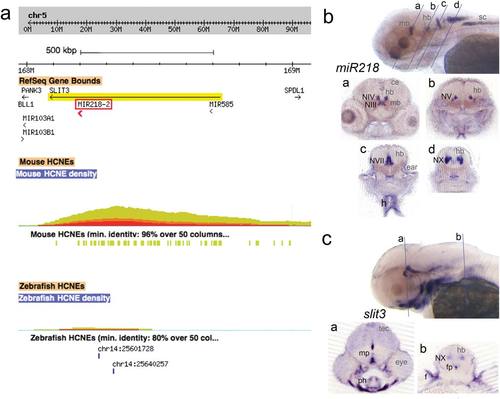
(a) Ancora Genome browser window of the human SLIT3 gene on chromosome 5, harboring miR218-2 in intron 14. The red-yellow colored hill represents the density of noncoding sequences that are conserved with the mouse genome (minimum identity 96% over 50 bp; illustrated as yellow bars). The highest density is ~100 kb distant of miR218-2. Conservation with the zebrafish genome is shown below. Only two HCNEs (80% identity over 50 bp) are present (illustrated by the blue bars). (b,c) miR218 LNA and slit3 in situ hybridization, shown are 4 dpf zebrafish larvae. Labeled structures are indicated unilaterally with black abbreviations. Abbreviations in grey are used to describe anatomical features. Abbreviations: ce, cerebellum; f, fin; fp, floorplate; h, heart; hb, hindbrain; mb, midbrain; mp, midbrain basal plate; NIII, oculomotor nerve nucleus; NIV, trochlear nerve nucleus; NV, trigeminal nerve nucleus; NVII, facial nerve nucleus; NX, vagal nerve nucleus; ph, pharyngeal arches skeleton; tec, tectum; vn, vagal nucleus. EXPRESSION / LABELING:
|
|
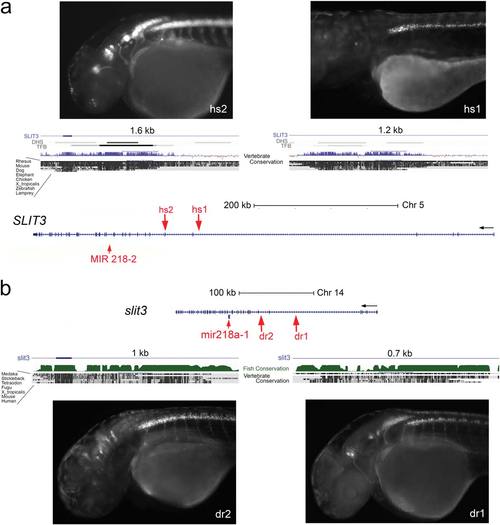
Elements amplified from human (hs) and zebrafish (dr) sequences within SLIT3/slit3 had specific enhancer activity that drove expression in a miR218-2 like pattern in brainstem- and spinal cord motor neurons. (a) Modified UCSC genome browser window showing the human miR218-2/SLIT3 locus. SLIT3 as well as the miRNA 218-2 are located in reverse orientation in the genome sequence. The 32 end of SLIT3 is to the left. miR-218-2 is located in the intron 14 of SLIT3. Two noncoding sequences conserved between human and zebrafish genomes were identified as enhancers of miR218-2. While hs2 drove GFP in most or allof the neuronal expression domain of miR218, hs1-driven GFP was seen only in spinal cord motor neurons. (b) In zebrafish the slit3 gene is much smaller than in the human genome. Two sequences that show conservation between human and zebrafish have enhancer activity in a pattern very similar to the enhancers identified in human SLIT3. Abbreviations: DHS, DNase 1 hypersensitive sites; TFB, transcription factor binding identified by ChIP seq – both tracks are provided by ENCODE. EXPRESSION / LABELING:
|
|
(a) zebrafish slit2 in situ hybridization. slit2 is expressed in the telencephalon, in the dorsal hindbrain, the midbrain basal plate and floorplate and the ganglion cell layer in the eye. Two sections, on the level of the hindbrain and of the spinal cord, show that cranial nerve nuclei and motor neurons are negative for slit2 transcripts. (b) Human SLIT2 gene with miRNA218-1 located in intron 14. The HS1 element (1165 bp) is 52 kb upstream including intron 5 and exon 6 sequences. UCSC genome browser indicates sequence conservation in all vertebrate genomes, including the basal vertebrate lamprey. (c) Transgenic larvae with hs1-driven GFP. The expression pattern is that of miR218, but when compared to the enhancers identified within SLIT3 (Fig. 2), the expression in the vagal nerve nucleus is much weaker and expression in anterior cranial nerve nuclei is missing. |
|
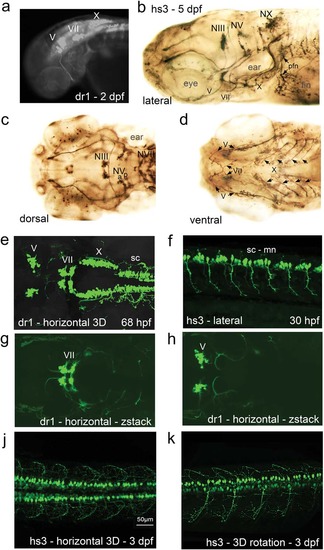
Two enhancers regulating zebrafish miR218a-1 (dr1) and human miR218-2 (hs3) drive GFP in transgenic embryos and larvae. GFP expression is found in cranial nerve nuclei and in spinal cord motor neurons and distributed in the axons. (a) Overview fluorescent microscope image. (b–d) GFP expression pattern was enhanced through anti-GFP immunostaining. The images are from different larvae and in different orientations. (e–k) Confocal live imaging. (e) z-stack image through the whole expression domain as scanned in this specimen. (f) Optical section showing spinal cord motor neurons in 30 hpf embryos. (g,h) Two z-stack images of the scan shown in “e”; one is further dorsal with focus on the facial nerve nucleus (g), while the trigeminal nerve nuclei appear further ventral in this scan (h). (j,k) 3D projections of spinal cord motor neurons. Abbreviations: hpf, hours post fertilization; mn, motor neurons; sc, spinal cord; NIII, oculomotor nerve nucleus; NV, trigeminal nerve nucleus (a, anterior; b, posterior); NVII, facial nerve nucleus; NX, vagal nerve nucleus; pfn, pectoral fin nerve. Note that V, VII, X are the corresponding nerves and X has several branches underneath the ear as seen in the lateral view. EXPRESSION / LABELING:
|