- Title
-
Phenotype-driven chemical screening in zebrafish for collective cell migration inhibitors identifies multiple potential pathways for targeting metastasis
- Authors
- Gallardo, V.E., Varshney, G.K., Lee, M., Bupp, S., Xu, L., Shinn, P., Crawford, N.P., Inglese, J., Burgess, S.M.
- Source
- Full text @ Dis. Model. Mech.
|
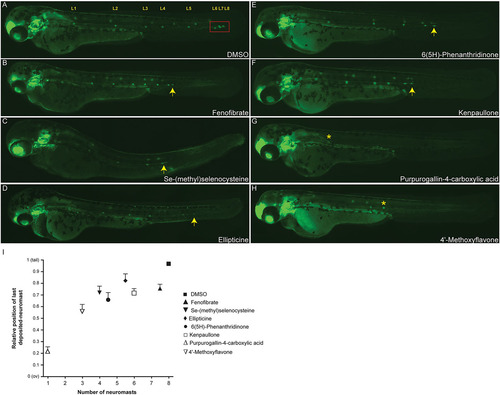
Examples of PLL migration phenotypes observed in the screen. Phenotypes of 48hpf zebrafish embryos treated with small molecules that had been identified in the screen. (A) DMSO control. The red box indicates the successful migration of the terminal neuromasts to the end of the tail. Cldnb:EGFP embryos treated with 10µM of Se-(methyl)selenocysteine (C), Ellipticine (D) or 6(5H)-Phenanthridinone (E) show a delay in primordium migration and, therefore, have disrupted lateral line formation compared with the DMSO control (A,I), whereas treatment with Fenofibrate (B) or Kenpaullone (F) causes an increase in the neuromast deposition rate, resulting in a phenotype with premature deposition of terminal neuromasts. Treatments with the flavonoid-derivative molecules Purpurogallin-4-carboxylic acid (G) and 42-Methoxyflavone (H), almost completely abolished the posterior lateral line formation, generating PLL phenotypes of 1 (G) and 3 (H) deposited neuromasts, respectively. Arrows indicate the position of the PLLp relative to the fully successful migration marked by the red box in A; asterisks indicate the position of the last deposited neuromast in G and H. (I) Quantification of the average position of the final deposited neuromast in drug-treated embryos relative to the distance from the otic vesicle (ov) to the tip of the tail. Experiments were carried out using 15 larvae per condition. Error bars indicate +s.d. Compounds showed a variety of effects, from inhibiting both neuromast numbers and migratory behavior (e.g. Purpurogallin-4-carboxylic acid) to only slowing migration (Fenofibrate). |
|
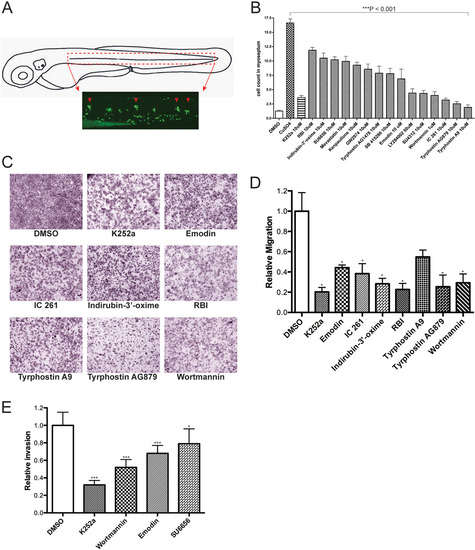
Confirmation of active compounds. (A) Schematic view of a 3day post-fertilization (dpf) larva. The boxed area corresponds to the horizontal myoseptum (dotted red lines) and shows the area where leukocytes were counted in all quantification experiments. (B) Quantification of infiltrating leukocytes in the lateral line after treatments with diverse inhibitors. The assays were carried out by manually counting leukocytes recruited to the lateral line neuromasts after Cu2+ treatment (10µM) as described in Results. In this experiment, drugs were added 30min prior to Cu2+ treatment. The graph shows average leukocyte numbers in the lateral line in negative controls (DMSO), in Cu2+ (as CuSO4) and in Cu2+-plus drug-treated fish. Leukocyte quantification shows significant reduction (***P<0.001) in neutrophil recruitment to damaged PLL neuromasts in larvae exposed to the compounds tested compared with Cu2+-exposed larvae, suggesting an inhibition of the neutrophil migration. Experiments were carried out with 15 larvae per condition. (C) Effects of kinase inhibitor compounds from LOPAC1280 on in vitro melanoma cell migration/invasion. 501-mel human melanoma cells were treated with DMSO (negative control) or the c-Met inhibitor K252a (positive control), and with the drugs indicated for 16h and then assayed for motility as described in Materials and Methods. Representative data from independent experiments are shown. Only compounds that had a negative effect on migration are shown. (D) Quantification of 501-mel cell migration assays. The migratory cells were counted and the results expressed as the mean number of migratory cells from two independent experiments. Measurements were calculated relative to control (DMSO). (E) Comparative effects of tested drugs on the cell invasion capacity of melanoma cells. 501-mel cells were treated with DMSO, K252a, Wortmannin, Emodin and SU6656 and then plated on Matrigel inserts in presence of HGF at 50ng/ml for 24h. The invasive cells were counted and the results expressed as the mean number of migratory cells in five random microscopic fields. Measurements were calculated relative to control (DMSO). Data collected were from two independent experiments. ***P<0.001; *0.01<P<0.05. EXPRESSION / LABELING:
PHENOTYPE:
|
|
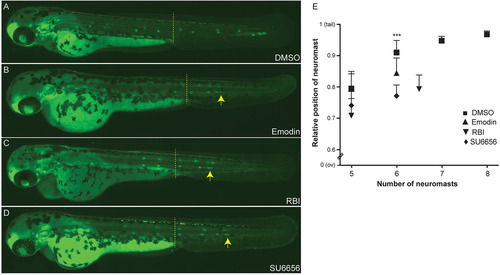
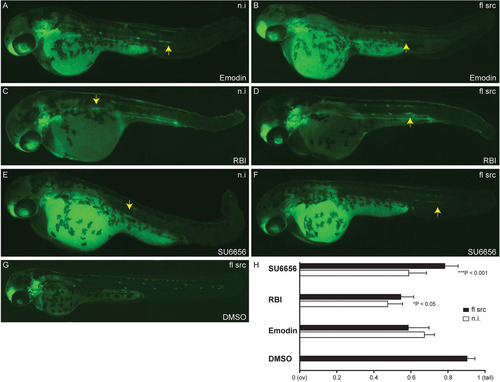
Blocking the Src signaling pathway inhibits collective cell migration in vivo. Cldnb:EGFP embryos were treated at 36hpf (primordium has migrated beyond the yolk extension, dashed line) with the c-Src inhibitors Emodin (B), RBI (C) or SU6656 (D). Treated embryos show a disruption of the primordium migratory ability compared with DMSO control (A), resulting in a phenotype with a premature deposition of a terminal neuromasts (L6) in all conditions (B-D). (E) Quantification of average position of the last five deposited neuromasts in drug treated-embryos relative to the distance from the ov to the tip of the tail. Arrows indicate the position of the PLLp. Experiments were carried out with ten larvae per condition. Error bars indicate +s.d.; ***P<0.001. |
|
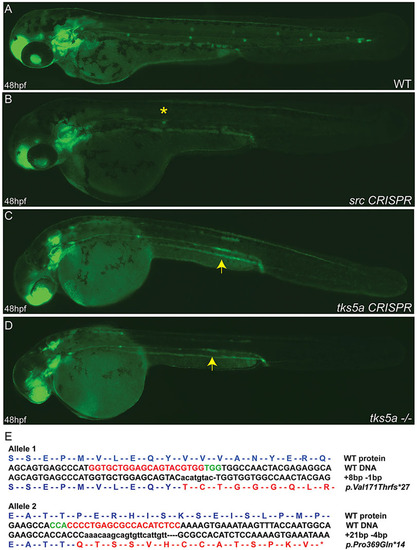
Disruption of src and tks5a genes by CRISPR/Cas9 affects the PLLp migration. Two sgRNAs targeting two different exons (25ng/µl) within each gene were co-injected with Cas9 mRNA (300ng/µl) into cldnb:EGFP embryos and the embryos raised. Injected fish were inbred and at 48hpf WT embryos show the normal pattern of the migrating primordium and neuromasts deposition (A), whereas src CRISPR (B), tks5a CRISPR (C) and tks5a/ (D) embryos show a disruption of the primordium migratory ability and PLL morphogenesis. Arrows indicate the position of the primordium in tks5a/ embryos; asterisk indicates the position of the last deposited neuromast. (E) Sequence confirmation of tks5a/alleles. Allele 1 has an 8-bp insertion and a 1-bp deletion, which changed amino acid valine at position 171 to threonine and truncates the protein after 27 amino acids. Allele 2 has a 21-bp insertion and a 4-bp deletion, which changed amino acid proline at position 369 to glutamine and truncated the protein after 14 amino acids. |
|
Overexpression of src mRNA rescues PLLp migration disrupted by Src signaling pathway inhibitors. Phenotypes of 48hpf cldnb:EGFP embryos upon overexpression of src followed by treatment with Src inhibitors that had been identified in the screen. Overexpression of full-length (fl) src in embryos following DMSO treatment show a normal lateral line development (G). Overexpression of src partially rescued primordium migration defects in embryos treated with RBI (D) and SU6656 (F) compared with non-injected-embryos (C,E), respectively. This phenotype was not rescued in Emodin-treated embryos upon overexpression of src (A,B). (H) Quantification of the average primordium position in drug-treated embryos upon src overexpression relative to the distance from the otic vesicle (ov) to the tip of the tail. Experiments were carried out with ten larvae per condition. Error bars indicate +s.d. ***P<0.001; *P<0.05. mRNA (100pg/2nl) was injected into one-cell stage embryos. n.i., non-injected-embryo. |
|
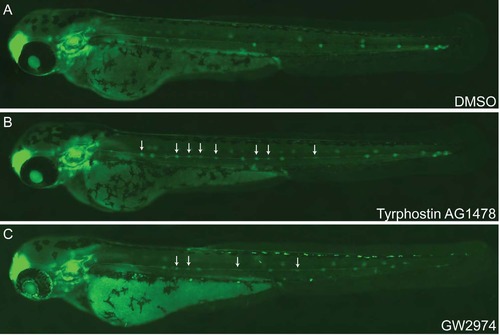
Regulation of neuromast number by ErbB signaling. Phenotypes of 48 hpf zebrafish embryos treated with ErbB signaling pathway inhibitors showed an unusual increase in neuromast numbers. Treatments of 20 hpf cldnb:EGFP embryos with 10µM of Tyrphostin AG1478 (B) and GW2974 (C) increased the number of deposited neuromasts compared to DMSO control (A). Arrow indicates the aberrant organization of interneuromast cells in treated embryos as compared to control. |
|
Supplemental figure 2. Chemically induced inflammation (ChIn) assay. DMSO-treated Tg(mpx:EGFP) larvae show the normal distribution of labeled cells, mostly localized in the ventral trunk and tail (A). In copper-treated siblings (B), leukocytes become localized preferentially to a few clusters along the horizontal midline of the trunk and tail (white arrows). Larvae exposed to indicated compounds (C-H) exhibit a decreasing number of leukocytes along the PLL. Inhibitors (C-H) were added to the incubation medium 30 min prior to addition of copper and were tested for inhibition of leukocyte migration using ChIn assays. |