- Title
-
Matricellular protein SPARC/Osteonectin expression is regulated by DNA methylation in its core promoter region
- Authors
- Torres-Núñez, E., Cal, L., Suarez-Bregua, P., Gomez-Marin, C., Moran, P., Gomez-Skarmeta, J., Rotllant, J.
- Source
- Full text @ Dev. Dyn.
|
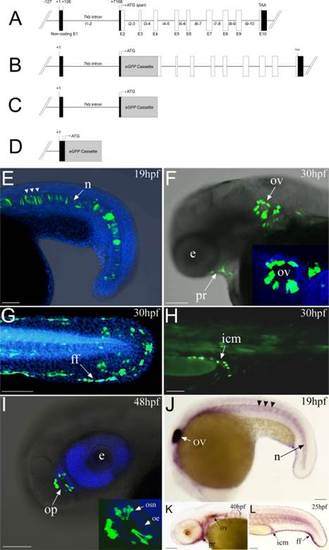
Sparc transgene constructs and the eGFP expression in zebrafish. A: A schematic diagram of the zebrafish sparc locus shows the exons and approximately 7,25 kb of the genomic region upstream of sparc. B: Sparc-iTol2-eGFP-BAC: eGFP modified BAC showing reporter eGFP gene inserted before sparc translational start codon. C: 7,25kb-sparc-Tol2-eGFP: Tol2 eGFP reporter gene construct contains a promoter region spanning nt 127 to the transcription start site (+1), the first exon, the unique intron of the 52-UTR of the gene and a small part of the second exon. D: 0,25kb-sparc-Tol2-eGFP is an eGFP reporter gene construct having a deletion of the noncoding intron 1–2 sequence. E–I: eGFP expression in zebrafish carring a modified BAC construct (Sparc-iTol2-eGFP-BAC) and/or a Tol2 eGFP reporter gene construct (7,25kb-sparc-Tol2-eGFP). Observed fluorescence from both construct was equivalent. Expression of eGFP(green) together with 4,6-diamidino-2-phenylindole (DAPI) (blue) was analyzed at the indicated developmental stages. E: At 19 hpf, periodic expression of eGFP is detected in the notochord (arrows). At 30 dpf, eGFP is expressed in a broad domain that contains most of the pharyngeal region (F), otic vesicle (F) (inset is a magnified image of the otic vesicle), fin fold (G) and intermediate cell mass (H). At 48hpf, eGFP is detected in the olfactory sensory neurons within the olfactory placode (I, inset is a magnified image of the olfactory placode, showing the olfactory sensory neurons and the olfactory epithelium). J–L: Whole mount in situ hybridization shows sparc mRNA expressed in zebrafish notochord (periodic expression, arrows), pharyngeal region, otic vesicle, find fold and hematopoietic ICM region at 19, 25, and 40 hpf stages. Lateral views with anterior to the left and dorsal to the top. Abbreviations: E, exon; i, intron; n, notochord; pr, pharyngeal region; ov, otic vesicle; ff, fin fold; e, eye;op, olfactory placode; osn, olfactory sensory neurons; oe, olfactory epithelium; icm, intermediate cell mass. Scale bars = 100 µm. Black boxes indicate untranslated regions; open boxes indicate coding exons; lollipops indicate CpG island detected. |
|
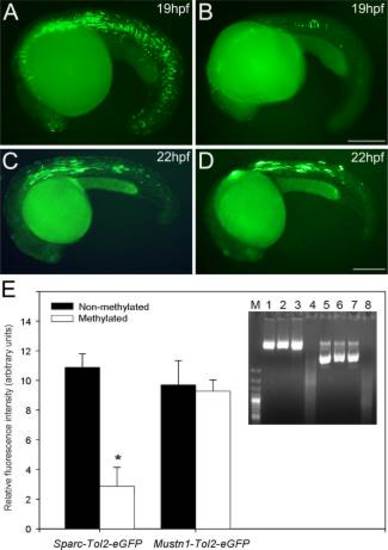
Effects of methylation on zebrafish sparc promoter activity in vitro. Zebrafish embryos were microinjected with 7,25kb-sparc-Tol2-eGFP or 0,7kb-Musnt1-Tol2-eGFP methylated (B,D) and unmethylated (A,C) promoter vectors. Microinjected methylated and unmethylated groups were as follows: (1) zebrafish 7,5Kb Sparc promoter cloned into pTolEGFPDest reporter plasmid (7,25kb-sparc-Tol2-eGFP) (A,B); (2) zebrafish 0,75Kb Mustnt1 promoter cloned into pTolEGFPDest reporter plasmid (0,7kb-Mustn1b-Tol2-eGFP) (Ctrl, CpG-free Promoter) (C,D). E: Intensity of eGFP fluorescent measurements were performed with Image J and displayed as vertical bar chart. Student t tests were performed to determine statistical relevance. Values are shown as mean ± S.E.M. (n = 10). E Inset: Successful vector methylation verification by analysis of band patterns on electrophoresis gel after digestion of the purified plasmids with the McrBC enzyme. Lane M, 1 Kb marker; lanes 1 and 5, 1 µg 7,25kb-sparc-Tol2-eGFP or 0,7kb-mustn1b-Tol2-eGFP; lanes 2 and 6, 1 µg 7,25kb-sparc-Tol2-eGF or 0,7kb-mustn1b-Tol2-eGFP treated with McrBC; lanes 3 and 7, 1 µg SssI-methylated 7,25kb-sparc-Tol2-eGFP or 0,7kb-mustn1b-Tol2-eGFP; lanes 4 and 8, 1 µg SssI-methylated 7,25kb-sparc-Tol2-eGFP or 0,7kb-mustn1b-Tol2-eGFP treated with McrBC. As expected, only the methylated vector was digested. Scale bars = 250 µm. |