- Title
-
Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- Authors
- Boer, E.F., Howell, E.D., Schilling, T.F., Jette, C.A., Stewart, R.A.
- Source
- Full text @ PLoS Genet.
|
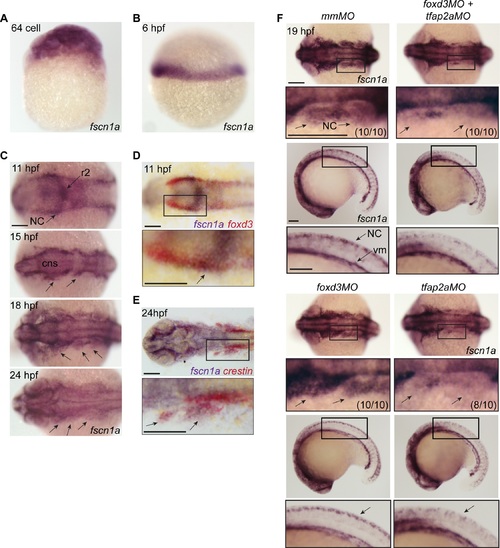
fscn1a mRNA expression in zebrafish NC is regulated by tfap2a. (A) Whole-mount ISH of maternal fscn1a mRNA at the 64-cell stage. (B) fscn1a mRNA expression at 6 hours post fertilization (hpf) at the gastrula margin. (C) Dorsal cranial views of fscn1a expression in 11, 15, 18, and 24 hpf embryos. 11 hpf embryos show expression in the brain, particularly rhombomere 2 (r2), and neural crest (NC) (top panel). In 15 and 18 hpf embryos, fscn1a is expressed in the brain (CNS) and cranial NC streams (middle panels, arrows). In 24 hpf embryos, fscn1a is expressed in the brain and pharyngeal arches (bottom panel, arrows). (D) In 11 hpf embryos, fscn1a mRNA is co-expressed with foxd3 at the neural plate border, highlighted in a magnified view (bottom panel). (E) fscn1a mRNA is co-expressed with crestin in cranial NC streams at 24 hpf, highlighted in a magnified view (bottom panel). (F) Dorsal cranial (top panels) and lateral (bottom panels) views of fscn1a expression in 19 hpf embryos injected with mismatch MO (mmMO), foxd3MO and tfap2aMO (foxd3MO+tfap2aMO), foxd3MO, or tfap2aMO. Magnified views of boxed regions are below each panel. Numbers in parentheses indicate proportion of embryos with phenotype. Scale bars = 100 µm. |
|
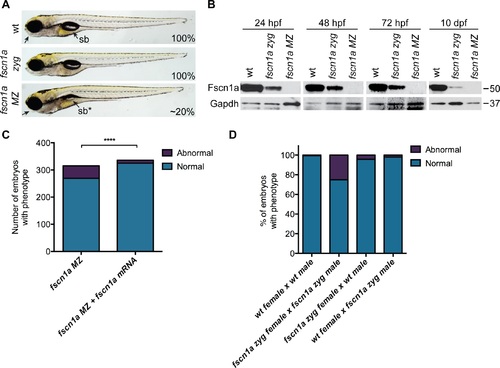
Maternal Fscn1a protein persists throughout embryogenesis and masks morphological defects in fscn1a zygotic mutants. (A) Brightfield images of 5 dpf wild type (wt), zygotic fscn1a (fscn1a zyg), and maternal/zygotic fscn1a (fscn1a MZ) embryos. Arrow highlights craniofacial cartilage, sb = swim bladder. Number indicates percentage of embryos with phenotype. (B) Immunoblot showing Fscn1a protein levels at 24, 48, 72 hpf and 10 dpf in wt, fscn1a zyg, and fscn1a MZ mutant embryos. Numbers on right denote molecular mass markers. (C) Quantitation of uninjected or fscn1a mRNA-injected fscn1a MZ embryos with normal vs. abnormal craniofacial cartilages at 5 dpf (n = 315 fscn1a MZ embryos, 336 fscn1a MZ + fscn1a mRNA embryos, ****p<0.0001). (D) Quantitation of embryos with normal vs. abnormal craniofacial cartilages at 5 dpf (n = 201 wt female × wt male, 119 fscn1a zyg female × fscn1a zyg male, 115 fscn1a zyg female × wt male, 151 wt female × fscn1a zyg male). PHENOTYPE:
|
|
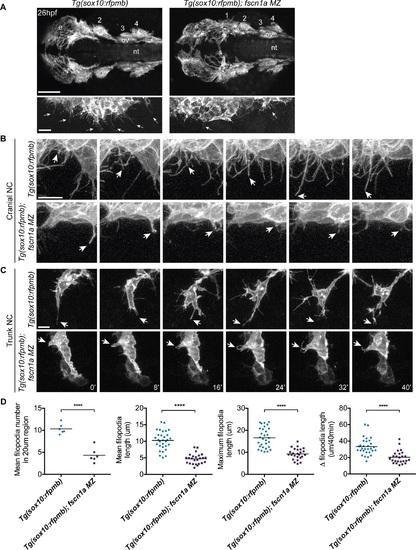
fscn1a is required for filopodia formation in NC cells. (A) Dorsal cranial views of cranial NC streams in 26 hpf Tg(sox10:rfpmb) and Tg(sox10:rfpmb); fscn1a MZ embryos, anterior is to the left. Numbers correspond to pharyngeal arches, e = eye, ov = otic vesicle, nt = neural tube, scale bar = 100µm. Leading edge of NC stream 1 is enlarged in right panel, scale bar = 10 µm. Arrows highlight filopodia at leading edge of NC stream 1. (B-C) Time lapse confocal images of filopodia at the leading edge of NC stream 2 (B) or of leading cells in trunk NC streams (C) in live Tg(sox10:rfpmb) and Tg(sox10:rfpmb); fscn1a MZ embryos at 26 hpf. Arrows highlight tips of single filopodia throughout the time lapses. Scale bar = 10 µm. (D) Quantitation of filopodia number, mean and maximum filopodia length, and change in filopodia length in Tg(sox10:rfpmb) and Tg(sox10:rfpmb); fscn1a MZ cranial NC (n = 6 embryos and 30 filopodia each for Tg(sox10:rfpmb) and Tg(sox10:rfpmb); fscn1a MZ, ****p<0.0001). |
|
fscn1a is required for collective migration of the first NC stream. Dorsal cranial views of wild type and fscn1a MZ embryos analyzed by RNA in situ hybridization for (A) foxd3 mRNA at 11 hpf, (B) sox10 mRNA at 15 hpf, showing asymmetric dispersion of NC cells in fscn1a MZ (arrow), and (C) dlx2a mRNA at 18, 28 and 36 hpf. Numbers correspond to pharyngeal arches, arrows indicate abnormal NC migration phenotypes observed in dlx2a+ NC cells in fscn1a MZ embryos. Anterior is left in all panels. Phenotypes shown in fscn1a MZ panels were observed in 100% (A) or ~20% (B-C) of embryos examined (n>200). EXPRESSION / LABELING:
PHENOTYPE:
|
|
fscn1a is required for development of NC-derived craniofacial cartilage. (A) Lateral views of wild type (wt) and fscn1a MZ 5 dpf embryos. Arrow indicates reduced jaw in fscn1a MZ embryos. (B) Lateral (left) and ventral (right) views of Alcian blue-stained wt and fscn1a MZ 5 dpf embryos. Numbers correspond to pharyngeal arches. Arrow indicates missing palatoquadrate in fscn1a MZ embryos. (C) Ventral view of dissected Alcian blue-stained pharyngeal skeletons from wt and fscn1a MZ 5 dpf embryos. ch = ceratohyal, cb = ceratobranchial, hm = hyomandibular, hs = hyosymplectic, mk = Meckel’s cartilage, pq = palatoquadrate. PHENOTYPE:
|
|
Depletion of fscn1a-independent NC cell filopodia by low concentration Latrunculin B treatment does not enhance the severity of fscn1a MZ craniofacial defects. (A) Lateral views of leading edge of cranial NC stream 1 in 26 hpf DMSO or Lat. B-treated Tg(sox10:rfpmb) or Tg(sox10:rfpmb); fscn1a MZ embryos. Scale bar = 25 µm. (B) Quantitation of filopodia number and filopodia length in DMSO or Lat. B-treated Tg(sox10:rfpmb) and Tg(sox10:rfpmb); fscn1a MZ embryos (n = 9 Tg(sox10:rfpmb) + DMSO, n = 10 Tg(sox10:rfpmb) + Lat. B, n = 10 Tg(sox10:rfpmb); fscn1a MZ + DMSO, n = 10 Tg(sox10:rfpmb); fscn1a MZ + Lat. B, *p = 0.02, **p<0.005, ***p<0.001, ****p<0.0001, ns = not significant). (C) Representative ventral views of 5 dpf DMSO or Lat. B-treated wild type (wt) and fscn1a MZ embryos stained with Alcian blue. (D) Quantitation of percentage of embryos with normal or abnormal craniofacial development at 5 dpf. Numbers above columns indicate number of embryos included in analysis (n = 83 wt + DMSO, 93 wt + Lat. B, n = 86 fscn1a MZ + DMSO, n = 72 fscn1a MZ + Lat. B, *p = 0.02). |
|
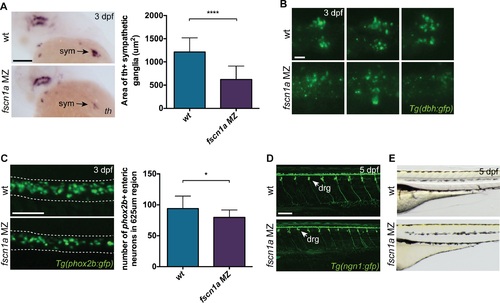
fscn1a is required for development of a subset of NC derivatives. (A) Lateral views of 3 dpf wt and fscn1a MZ embryos stained for th. sym = sympathetic ganglia, scale bar = 100 µm. Mean area of th+ sympathetic ganglia is plotted in graph on right. (n = 25 wt, 24 fscn1a MZ, ****p<0.0001). (B) Representative images of dbh:gfp+ sympathetic ganglia in 3 dpf wt and fscn1a MZ embryos. Dorsal views, anterior to the left. Scale bar = 25 µm. (C) Representative images of phox2b:gfp+ enteric neurons in 3 dpf wt and fscn1a MZ embryos. Dotted lines outline gut. Scale bar = 100 µm. Number of phox2b:gfp+ enteric neurons in 625 µm region of gut is plotted in graph on right (n = 15 wt, 13 fscn1 MZ, *p = 0.03). (D) ngn1:gfp+ dorsal root ganglia (drg) in 5 dpf wt and fscn1a MZ embryos. Scale bar = 100 µm. (E) Lateral views showing melanophores in 5 dpf wt and fscn1a MZ embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
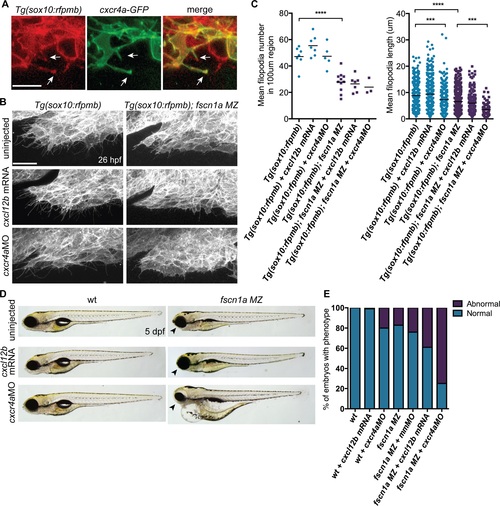
fscn1a cooperates with cxcr4a/cxcl12b signaling to regulate directional cranial NC migration. (A) Representative confocal images of cranial NC cells from a 26 hpf Tg(sox10:rfpmb) embryo injected with cxcr4a-GFP mRNA. Arrows highlight Cxcr4a-GFP localized to NC filopodia. Scale bar = 10 µm. (B) Lateral view of leading edge of cranial NC stream 1 in 26 hpf uninjected, cxcl12b mRNA-injected or cxcr4aMO-injected Tg(sox10:rfpmb) or Tg(sox10:rfpmb); fscn1a MZ embryos. Scale bar = 25 µm. (C) Quantitation of filopodia number and filopodia length in uninjected, cxcl12b mRNA-injected or cxcr4aMO-injected Tg(sox10:rfpmb) and Tg(sox10:rfpmb); fscn1a MZ embryos (n = 7 Tg(sox10:rfpmb), n = 9 Tg(sox10:rfpmb) + cxcl12b mRNA, n = 5 Tg(sox10:rfpmb) + cxcr4aMO, n = 7 Tg(sox10:rfpmb); fscn1a MZ, n = 8 Tg(sox10:rfpmb); fscn1a MZ + cxcl12b mRNA, n = 3 Tg(sox10:rfpmb); fscn1a MZ + cxcr4aMO, ***p<0.001, ****p<0.0001). (D) Lateral views of 5 dpf uninjected, cxcl12b mRNA-injected or cxcr4aMO-injected wild type (wt) and fscn1a MZ embryos. Arrowheads indicate reduced jaw. (E) Quantitation of percentage of embryos with normal or abnormal craniofacial development at 5 dpf (n = 43 wt, 104 wt + cxcl12b mRNA, n = 51 wt + cxcr4aMO, n = 165 fscn1a MZ, n = 130 fscn1a MZ + mmMO, n = 123 fscn1a MZ + cxcl12b mRNA, n = 154 fscn1a MZ + cxcr4aMO). |
|
Expression of fscn1b during embryonic development. (A) Whole-mount ISH for fscn1b mRNA expression at 11 hpf (3 somite), 18 hpf (18 somite), 24 hpf, 36 hpf, and 48 hpf embryos. fscn1b expression is observed in specific neurons in the developing brain at 36 hpf and 48 hpf (arrows). (B) Whole-mount ISH for fscn1b mRNA expression in 36 hpf embryos using a control sense RNA probe. Black pigment in eyes, trunk and over yolk is visible. EXPRESSION / LABELING:
|
|
Expression of fscn1a in trunk NC. (A) Lateral views of fscn1a or crestin mRNA expression in 18 hpf embryos. Trunk is enlarged in lower panels. Arrows highlight NC expression. Scale bar = 100 µm. (B) Lateral views of fscn1a mRNA expression in a 24 hpf embryo (top panel) and with crestin (bottom panels) showing co-expression in NC but not vasculogenic mesoderm (vm). Boxed region is enlarged in right panel. Scale bar = 100 µm. (C) Whole-mount ISH for fscn1a mRNA expression in 24 hpf embryos using a control sense RNA probe. EXPRESSION / LABELING:
|
|
Loss of fscn paralogs does not enhance fscn1a MZ craniofacial defects. Ventral views of 5 dpf uninjected or fscn1b/2a/2bMO-injected wild type and fscn1a MZ embryos stained with Alcian blue. Numbers in parentheses indicate number of embryos with depicted phenotype. |
|
fscn1a MZ embryos display abnormal craniofacial skeleton morphology. Representative images of craniofacial skeleton phenotypes observed in fscn1a MZ embryos. Ventral views of craniofacial skeleton in 5 dpf Tg(sox10:rfpmb); fscn1a MZ embryos. Phenotypes are grouped into three classes: normal craniofacial cartilage morphology (Class I), mild deformation of mandibular arch (Class II) and loss of mandibular arch elements (Class III). Within a clutch of fscn1a MZ embryos, ~80% of embryos belong to Class I or II. The remaining 20% of embryos display symmetric or asymmetric loss of mandibular arch elements of variable severity and belong to Class III. Numbers denote pharyngeal arches, ma = mandibular arch (arch 1), ep = ethmoid plate. Scale bar = 100 µm. |
|
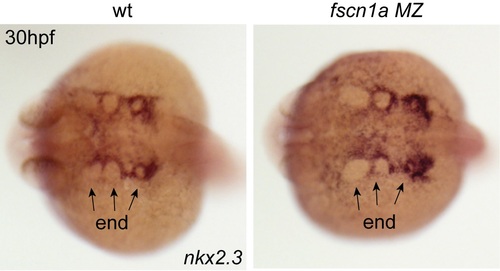
nkx2.3+ pharyngeal endoderm is patterned normally in fscn1a MZ embryos. Whole-mount in situ hybridyzation for nkx2.3 in wild type and fscn1a MZ embryos at 30 hpf showing normal endodermal (end) pouch formation. EXPRESSION / LABELING:
|
|
fscn1a cooperates with cxcr4a/cxcl12b signaling to regulate development of a subset of NC derivatives. (A) Representative ventral views of 5 dpf uninjected or cxcl12b mRNA-injected wild type and fscn1a MZ embryos stained with Alcian blue. (B) Whole-mount ISH for th expression in 4 dpf uninjected or cxcr4aMO-injected wt and fscn1a MZ embryos. (C) Average area of th+ sympathetic ganglia (n = 8 wt, 15 wt + cxcr4aMO, 17 fscn1a MZ, 17 fscn1a MZ + cxcr4aMO, **p = 0.0039, ***p = 0.0003, ****p<0.0001, sym = sympathetic ganglia). |
|
Low dose Latrunculin B treatment inhibits F-actin polymerization in residual NC cell filopodia. Representative maximum projection confocal images of the leading edge of second cranial NC stream from 26 hpf Tg(sox10:rfpmb) embryos injected with lifeact-GFP mRNA and treated with low dose Lat. B or DMSO. Arrows indicate filopodia. Asterisk denotes lack of Lifeact-GFP in RFP+ filopodia in Lat. B-treated embryo. F-actin in underlying yolk cells is also visible in all images. |