- Title
-
Simplet/Fam53b is required for Wnt signal transduction by regulating β-catenin nuclear localization
- Authors
- Kizil, C., Küchler, B., Yan, J.J., Özhan, G., Moro, E., Argenton, F., Brand, M., Weidinger, G., Antos, C.L.
- Source
- Full text @ Development
|
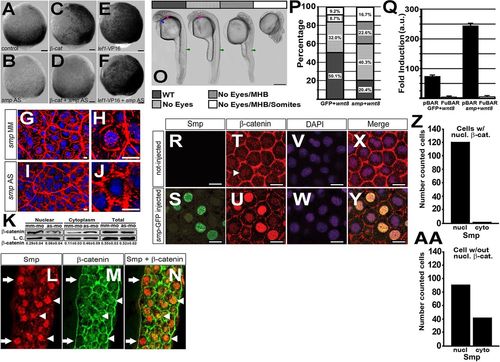
Loss of smp causes Wnt-related developmental phenotype and loss of Wnt-dependent gene expression. (A) smp mRNA (dark purple) in the single-cell embryo. (B,C) The smp transcript in 32-cell embryo (B) and in 1000-cell embryo (C). Expression remains ubiquitous at 50% epiboly (D) and at tailbud (lateral view) stage (E). (F) Control embryo (MM) 24h post-fertilization (hpf) after injection with mismatch smp morpholino at the one-cell stage. In situ hybridization with a xirp2 probe highlights somatic muscle boundaries. (G,H) Injection of antisense morpholino to the start (ATG) site (G) or first intron-exon (splice) boundary (H) of smp in 24hpf embryos. Activation of 7xTCF:mCherry in mismatch control embryos (I,K,M) compared with smp morphants (J,L,N). Activity of Top:dGFP in control embryos (O,Q) compared with smp morphants (P,R). cdx4 expression at indicated stages in mismatch controls (S,U) and in smp morphants (T,V). Expression of tbx6 at indicated stages in controls (W,Y) and smp morphants (X,Z). Expression of gbx1 at tailbud stage in controls (AA) and smp morphants (BB). Numbers in lower right corners of the panels indicate the number of embryos with the depicted expression pattern/the total number of animals. Scale bars: 100μm. |
|
Smp regulates β-catenin nuclear localization. (A) Activity of the 7xTCF:mCherry transgene at 95% epiboly after mismatch morpholino injection, (B) knockdown of smp, (C) overexpression of β-catenin in mismatch controls and (D) overexpression of β-catenin in smp morphants. (E) Activity of the 7xTCF:mCherry transgene at 95% epiboly after overexpression of lef1-VP16 in mismatch controls and (F) overexpression of lef1-VP16 in smp morphants. (G,H) Immunohistochemistry staining for β-catenin (red) and staining of nuclei with DAPI (blue) in mismatch control and (I,J) smp morphants. (K) Western blots for β-catenin in nuclear and cytoplasmic lysates from mismatch-morpholino controls (mm-mo) and smp morphants (as-mo). Total amount of β-catenin levels in the cells was unaltered by the smp knockdown. Loading controls were γ-tubulin for the cytoplasmic fraction and H2A for the nuclear fraction. (L) Immunohistochemistry staining for zebrafish Smp shows positive nuclei in the marginal zone where Wnt signaling is active. (M) Immunohistochemistry staining for β-catenin shows localization at the plasma membrane and in distinct nuclei (white arrowheads). (N) Merged stainings show colocalization of Smp and β-catenin in nuclei (white arrowheads) and cells with Smp in nuclei lacking β-catenin (white arrows). (O) Overexpression of wnt8 during zebrafish development produces phenotype classes that affect normal development of eyes (blue arrow); the midbrain-hindbrain boundary (pink arrow); and the somites and posterior structures (green arrows). (P) Percentage occurrence of phenotypic classes produced by overexpression of wnt8 alone or overexpression of wnt8 with smp. (Q) Results of luciferase assays of either the pBAR reporter (β-catenin binding sites) or the pFuBAR reporter (mutated β-catenin sites) for Wnt activity in HEK293T cells. (R) Uninjected controls. (S) GFP localization in the nuclei of embryos injected with mRNA encoding Smp-GFP. (T) β-Catenin localization in the dorsal region of control embryos. (U) β-Catenin localization in Smp-GFP-injected embryos. (V,W) DAPI staining labels nuclei. (X,Y) Merged fluorescence for β-catenin, GFP and DAPI. (Z) The number of β-catenin-positive cells with Smp in the nucleus or in the cytoplasm. (AA) The subcellular distribution of Smp in cells lacking β-catenin nuclear staining. Scale bars: 10μm in A-G,I,L-N; 1μm in H,J; 300μm in O; 10μm in R-Y. All experiments were performed at least three times. Data represent the mean; error bars indicate s.d. EXPRESSION / LABELING:
|
|
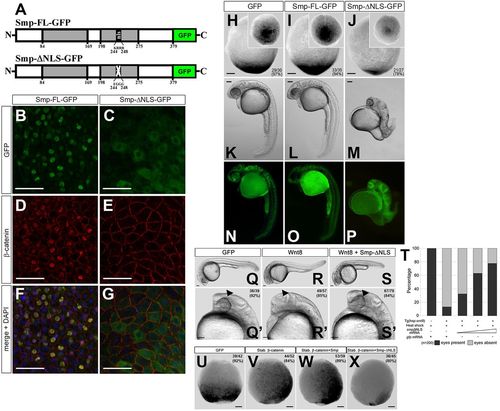
Nuclear localization of β-catenin requires the Smp nuclear localization signal. (A) The domain structure of Smp. The two grey boxes indicate regions of significant conservation among vertebrates. The numbers indicate the position of the amino acids. ‘nls’ identifies the nuclear localization signal KRRR. Substitution of these amino acids with EGGG ablates the nuclear localization signal (white cross). (B) Overexpression of Smp-GFP in fish embryos. (C) Deletion of the nuclear localization signal in Smp-GFP. (D) Co-staining for β-catenin in Smp-GFP-injected embryo. (E) Co-staining for β-catenin in Smp-ΔNLS-GFP-injected embryo. (F) Overlay of Smp-GFP and β-catenin immunostainings in Smp-GFP-injected embryo. (G) Overlay of Smp-GFP and &neta;-catenin immunostaining in Smp-ΔNLS-GFP-injected embryo. (H-J) Overexpression of GFP (H), Smp-FL (I) or Smp-ΔNLS (J) in the Tg(7xTCF:mCherry) embryos at tailbud stage. (K) Overexpression of GFP in a 24hpf embryos. (L) Overexpression of Smp-GFP in 24hpf embryo. (M) Overexpression of Smp-ΔNLS-GFP in 24dpf embryo. (N-P) Expression of each GFP-fused construct in the injected embryos. (Q,Q2) Control-injected transgenic embryos at 24hpf. (R,R′) Transgenic overexpression of wnt8. (S,S′) Injection of smp-ΔNLS into embryos transgenically overexpressing wnt8. Arrowheads indicate the expected location of the developing eye. (T) Graph shows a direct correlation between the rescue from the wnt8-overexpression posteriorization phenotype and the amount of smp-ΔNLS mRNA injected. Data are the average percent. (U) Control-injected Tg(7xTCF:mCherry) embryos. (V) Injection of mRNA encoding stabilized β-catenin. (W) Co-injection of mRNAs for stabilized β-catenin and Smp-FL. (X) Overexpression of Smp-ΔNLS with stabilized β-catenin. Numbers in the lower or upper right panel corners represent number of embryos with the observed phenotype/the total number of embryos (also represented as a percentage). Scale bars: 50μm in B-G; 100μm in H-X. Numbers in the lower or upper right corners indicate the number of embryos with the depicted expression patterns/the total number of embryos. |
|
Mammalian SMP (FAM53B) regulates β-catenin similarly to zebrafish Smp and interacts with β-catenin to retain it in the nucleus. (A) Immunocytochemistry (ICC) for the human SMP (green) in the Hoechst-stained nuclei (blue) of HEK293T cells. (B) Higher magnification shows the protein localizes as foci in Hoechst-stained nuclei. (C-F) β-Catenin ICC (green) of control siRNA-transfected unstimulated HEK293T cells (C), SMP siRNA-transfected HEK293T cells (D). control siRNA-transfected HEK293T cells stimulated with Wnt3-conditioned medium (E) and SMP siRNA-transfected HEK293T cells stimulated with Wnt3-conditioned medium (F). (C′-F′) β-Catenin ICC merged with DAPI (blue). (G) Representative subcellular fractionation blots showing the protein levels of nuclear (Nucl.) and cytoplasmic (Cytopl.) β-Catenin in unstimulated and Wnt3-stimulated HEK293T cells transfected either with control (Ctr) siRNA or Smp siRNA. Fract. denotes nuclear or cytoplasmic fractionation blots. Lys. denotes blots for loading control of total lysates for each fraction. Laminin and GAPDH show clear separation of the fractions. The numbers indicate protein ladder positions in kDa. (H) Fold activation of pBAR luciferase reporter from assays of control and Smp siRNA knockdown HEK293T cells without and with Wnt3-condition medium stimulation (Wnt). P<0.01 between Ct and SMP siRNA Wnt3-stimulated groups. Data are the average±s.d. (I) Dose-dependent activation of β-catenin-responsive promoter by human SMP. *P<0.05, **P<0.001, ***P<0.008. Data are the average±s.d. (J) Immunoprecipitation blot using anti-Myc antibody to pull down Myc-tagged human SMP shows co-immunoprecipitation of Flag-tagged human β-catenin as detected by anti-Flag antibody. (K) Immunoprecipitation experiment using different TAP-Tagged (TAP) SMP deletion mutants to determine which conserved domain interacts with FLAG-tagged β-catenin. (L) Immunoprecipitation experiment of SMP-myc for different FLAG-tagged β-catenin deletion mutants lacking either the N terminus or different sets of armadillo repeats. The lysate gels show the level of expression of each construct. (M) Luciferase assay of HEK293T cells transfected with: pBAR and stabilized β-catenin; pBAR, stabilized β-catenin and full-length SMP; or pBAR, stabilized β-catenin and SMP lacking the first homology domain (SMP-ΔHRI). P<0.01 between all groups. Data are the average±s.d. (N) Transfection of HEK293T cells with TAP-tagged GFP or SMP-GFP or SMP-ΔHRI-GFP shows nuclear localization of both SMP expression constructs. (O) Blot of subcellular fractionation (nuclear versus cytoplasmic) experiment for transfected TAP-tagged Smp and FLAG-tagged β-catenin. Immunoprecipitation blots (IP) are above. Lysate blots (Lys) are below. Lysate blot probed with GAPDH and laminin shows clear separation of each fraction. (P) β-Catenin-mCherry fluorescence before bleaching and at increasing times after photobleaching the cytoplasm of HEK293T cells co-transfected either with GFP or SMP-GFP. The graph shows the decrease in nuclear β-catenin-mCherry and its cytoplasmic recovery. *P<0.05. Scale bars: 20μm in A,B; 5μm in C-F2′. Data represent at least three or more independent experiments. |
|
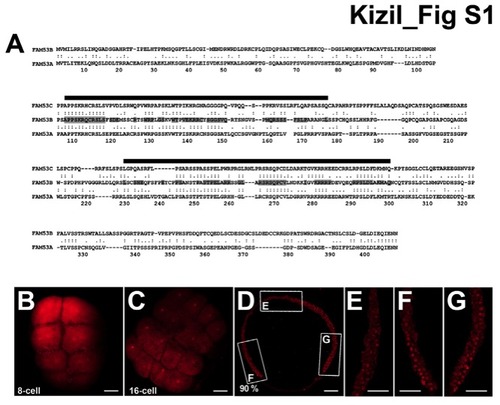
smp expression during embryonic development of zebrafish, Related to Figure 1. (A) Sequence comparison between human SMP/FAM53B and FAM53A and C, genes most closely related to Smp/Fam53b: sequence identities are 31% between B and A and 33% between B and C as shown by the conserved amino acids highlighted grey. The black lines indicate the two homology regions in the SMP/FAM53B protein described in this paper. (B) Immunohistochemistry staining for zebrafish Smp shows the presence of the maternal protein in the 8-cell embryo. (C) Immunohistochemistry for Smp at 16-cell stage also detects the presence of the protein. (D) Immunohistochemistry for Smp at 90% epiboly. Lettered boxes indicate location of panels E-G. (E-G) Enlarged images of anterior (E), ventral (F) and dorsal (G) sides of a stained zebrafish embryo at 90% epiboly shows broad expression of Smp protein primarily in the nuclei. Scale bars equal 50 μm (A-D) and 15 μm (E-G). |
|
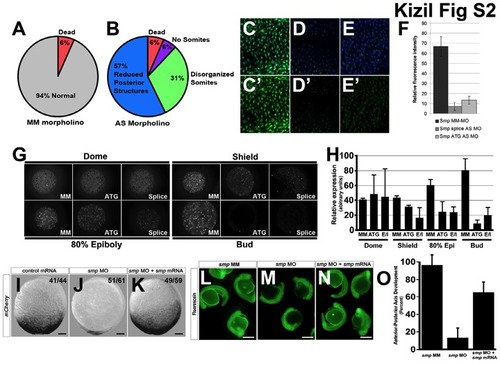
Characterisation of smp morpholinos, Related to Figure 1.(A) 94% of the embryos injected with the mismatch morpholino control for smp did not show a noticeable phenotype. (B) Knockdown of smp produced a majority of embryos lacking posterior structures, 31% that showed somites of different sizes, 6% that partially developed up to the start of somitogenesis and 6% that dead before somitogenesis. (C-E) Immunohistochemistry for zebrafish Smp in zebrafish embryos using the rabbit anti-zebrafish Smp (zSmp) polyclonal antibody and DAPI staining on mismatch smp control injected (C), antisense smp ATG morphant (D) and antisense smp splice site morphant embryos (E). (C′-D′) Immunostainings without DAPI staining. (F) Graph of the measurements of the staining levels of staining by the rabbit anti-zSmp polyclonal antibody of each injected morpholino group. (G) Expression of maternally loaded protein at different stages of development and effect on protein expression after morpholino knockdown (MM) Mismatch, (ATG) ATG morpholino, (Splice) Splice-site morpholino. (H) Quantitation of maternal protein expression profiles. (I) 7xTCF-mCherry activity at 95% epiboly after injection of GFP mRNA in the one-cell embryo. (J) 7xTCF-XLa.Siam:nlsmCherry activity in smp morphant. (K) 7xTCFXLa. Siam:nlsmCherry activity after co-injection of smp mRNA with smp morpholino. (L) 24 hour zebrafish embryos with fluorescein-labeled mismatch morpholino for smp show no developmental phenotype. (M) Knockdown of smp with a fluorescein-labeled morpholino shows severe axis anterior-posterior axis defects. (N) Rescue of knockdown phenotype when smp mRNA is co-injected with smp antisense morpholino. (O) Graph depicts quantitative assessment of the anterior-posterior axis phenotype by smp mRNA. Numbers in the lower left corners in panels (I-K) indicate the number of embryos with the depicted expression patterns to the total number of embryos. Scale bars are 50 μm (I-K) and 300 μm (L-N). |
|
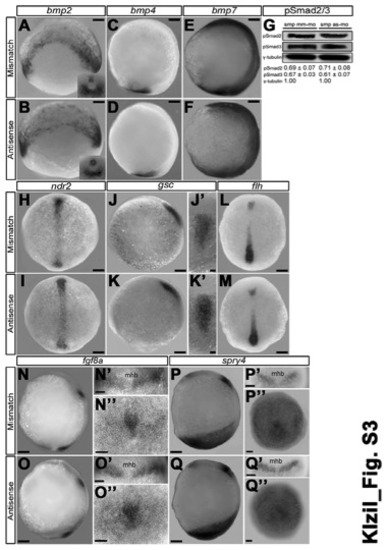
Loss of smp activity does not affect Bmp, Nodal or Fgf signaling, Related to Figures 1,4. (A) bmp2 expression in the marginal cells and the tailbud (insert) of mismatchmorpholino controls. (B) bmp2 expression in smp morphants (Antisense) shows similar expression as controls at tailbud stage. (C) bmp4 at the tailbud stage is located in the posterior tailbud. (D) bmp2 expression in smp morphants is similar to controls. (E) bmp7 expressed throughout the tailbud-staged embryo of mismatch controls. (F) smp morphants show similar bmp7 expression. (G) qRT-PCR comparing the relative levels of Np63 expression between MM control and smp morphants. (H) Dorsal view of ndr2 (nodal related 2) expression in the prechordal plate of control embryos at tailbud stage. (I) Dorsal view ndr2 expression in smp morphants. (J) Lateral view of goosecoid (gsc) expression in controls at tailbud stage. (J′) Dorsal view of gsc in controls. (K) Lateral view of gsc expression in smp morphants. (K′) Dorsal view of gsc expression in smp morphants. (L) Dorsal view of floating head (flh) expression the prechordal plate of controls. (M) Dorsal view of flh expression in smp morphants. (N) Lateral view of fgf8a expression in controls show expression in midbrainhindbrain boudary, and the mesendoderm of the tailbud. (N′) Dorsal view of fgf8a expression in midbrain-hindbrain of controls. (N′′) Posterior view of fgf8a expression in tailbud of controls. (O) Lateral view of fgf8a expression in smp morphants. (O′) Dorsal view of fgf8a expression in smp morphants. (O′′) Posterior view of fgf8a expression in smp morphants. (P) Lateral view of spry4 expression in controls showing midbrain-hindbrain expression and posterior expression. (P′) Dorsal view of spry4 expression in midbrain-hindbrain of controls. (P′′) Posterior view of spry4 expression in controls. (Q) Lateral view of spry4 expression in smp morphants. (Q′) Dorsal view of spry4 expression smp morphants. (Q′′) Posterior view of spry4 expression in smp morphants. Lateral views: A-F, J, K, N-Q; elsewhere dorsal (H, I, L, M), anterior (N′-Q′) or tailbud (N′′-Q′′) views. Numbers in the lower left corners in panels indicate the number of embryos with the depicted expression patterns to the total number of embryos. Scale bars: 100 μm (A-K,L-N,O,P,Q) and 50 μm elsewhere. Numbers below the Western blot bands indicate relative normalized abundance values. |
|
Altered cyp26a1 expression in smp morphants, Related to Figure 1. (A) raldh2 expression in the paraxial mesoderm of mismatch controls at tailbud (A) and 2-somite stage (A′) embryos. (B) raldh2 expression in smp morphants at tailbud (B) and 2-somite stage (B′). (C) cyp26a1 in the margin and the presumptive brain of controls at 95% epiboly. (D) smp morphants have similar expression at the same stage. (E) At tailbud stage, controls had cyp26a1 expression in the anterior pole. (F) smp morphants showed expansion of cyp26a1 expression. (G) cyp26a1 expression (blue) and 7xTCF-XLa.Siam:nlsmCherry reporter expression (red) in controls at 95% epiboly. (H) cyp26a1 expression does not change at 95% epiboly in antisense-morphant embryos, while mCherry expression (Wnt signaling) is reduced. (I) cyp26a1 expression at 95% epiboly in non-transgenic embryos after heat shock at 60% epiboly. (J) Expanded expression of cyp26a1 at 95% epiboly in embryos harboring the hsp70:dkk-1-GFP transgene after heat shock at 60% epiboly. Numbers in the lower left corners in panels indicate the number of embryos with the depicted expression patterns to the total number of embryos. Scale bars: 100 μm. |
|
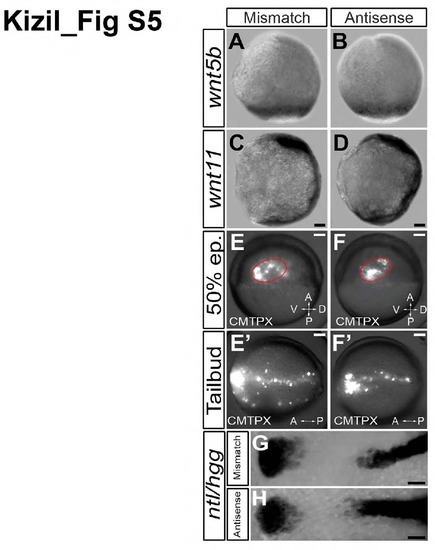
Convergent extension movements are not affected by knockdown of smp, Related to Figure 1. (A) wnt5b expression in the marginal zone of mismatch control embryos. (B) Similar expression of wnt5b in smp morphant (antisense) embryos. (C) wnt11 expression in the body axis of mismatch controls. (D) smp morphants show similar expression of wnt11 in the body axis. (E) Mismatch-control embryos were injected with CMTPX cell tracker dye into the upper marginal zone at 50% epiboly as shown by lateral view. (E′) Dorsal view of a mismatch-control embryo showing labeled cells throughout the embryonic body axis. (F) smp morphant embryos (lateral view) were similarly injected with CMTPX cell tracker dye at 50% epiboly. (F′) Dorsal view of CMTPX-positive cells align at the midline and cluster at the anterior region at tailbud stage similar to the mismatch control embryos. (G) Expression of no tail (ntl) (black arrows) in the posterior and calpthesin L (hgg) (white arrows) in the anterior of the mismatch control embryos. (H) Expression of ntl and hgg in smp morphants is similar to controls. Scale bars in A-F′ are 100 μm. |
|
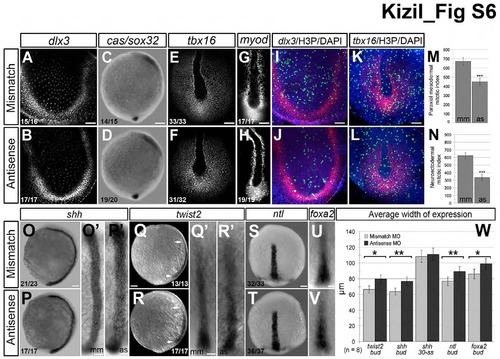
Gastrulation markers at tailbud stage are unaffected by loss of smp, Related to Figure 1. (A) Mismatch-morpholino-control embryos show dlx3b expression in the neuroectoderm. (B) dlx3b expression is similar in smp morphants (Antisense). (C) cassanova/sox32 (cas/sox32) is observed in developing endoderm in mismatch controls. (D) smp morphants display similar (cas/sox32) expression. (E) tbx16 expression in the axial and paraxial mesoderm of control embryos. (F) smp morphants display similar tbx16 expression. (G) myod staining in control embryos shows expression in the adaxial of the paraxial mesoderm. (H) smp morphants show similar myod expression. (I) Double staining for H3P and dlx3b. (J) Double staining for H3P and dlx3b in smp morphants show decrease in the number of H3P and dlx3b expression. (K) Double staining for H3P and tbx16 expression in controls. (L) Double staining for H3P and tbx16 in smp morphants. (M) Graph of the number of H3P-positive cells overlapping with confocal planes containing dlx3b expression. (N) Graph showing number of H3P-positive cells that overlap with confocal planes showing tbx16 expression. (O) Lateral view of shh expression in the developing prechordal plate of controls. (O′) Dorsal view of shh expression in control embryos. (P) Lateral view of shh expression in smp morphants show expression along the length of the AP axis. (P′) Dorsal view of shh expression in smp morphants shows that that its expression is wider than shh expression controls. (Q) Lateral view of twist2 expression in the developing prechordal mesoderm in controls. (Q′) Dorsal view of twist2 expression in prechordal plate of control embryos. (R) twist2 expression in smp morphants. (R′) Dorsal view of twist2 in smp morphants. (S) Dorsal view of early ntl expression in the developing prechordal plate. (T) Dorsal view of ntl expression in smp morphants. (U) Dorsal view of expression of foxa2 in prechordal plate of controls. (V) Dorsal view of foxa2 expression in smp morphants. (W) Measurements of the lateral dimensions of the prechordal plate genes show slightly increased dimensions of the prechordal plate in smp morphants and that this alteration is temporary based on measurements of the dimensions of shh expression at 5 somite embryos. Numbers in the lower left corners in panels indicate the number of embryos with the depicted expression patterns to the total number of embryos. Scale bars: 50 μm (A,B,E,F,I-L), 100 μm (C,D,O-T), 250 μm (G, H, O′-R′,U′,V′). |
|
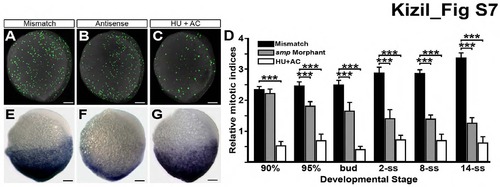
Loss of Wnt activity not due to defects in cell proliferation, Related to Figure 1. (A) Immunostaining for histone-3 phosphorylation (H3P) in MM-control embryos (mismatch) at 95% epiboly. (B) H3P staining is reduced in same-staged smp morphants (antisense). (C) H3P staining is decreased in embryos treated with hydroxyurea (HU) and aphidicolin (AC). (D) Graph indicates the relative mitotic indices of mismatch controls (black), smp morphant (grey) and HU/AC-treated embryos (white) through development. (E) 7xTCF-siam:mCherry activity in control embryos. (F) Activity of the Wnt-reporter in smp morphants. (G) Wnt-reporter activity in HU/AC-treated embryos. Scale bars equal 100 μm. “***” is p < 0.005. |
|
Apoptosis is not responsible for the loss of posterior structures in smp morphants, Related to Figure 1. (A) Measurement of TUNEL-positive cells in mismatch (MM-MO) control embryos and smp morphants (AS-MO). (B) Measurement of Annexin-Vpositive cells in MM-control embryos and smp morphants. (C) Graph showing the effect of p53 morpholino knockdown in MM-control and and smp-morphant embryos. (D) A 10-hour old p53-knockdown larva shows no effect on the expression of the 7xTCF:mCherry reporter. (E) smp-p53 double knockdown shows that loss of p53 does not restore the expression of the Wnt-dependent reporter in embryos lacking smp. (F) A smp morphant at 10 hours post fertilization shows loss of report expression. Scale bars equal 100 μm. |
|
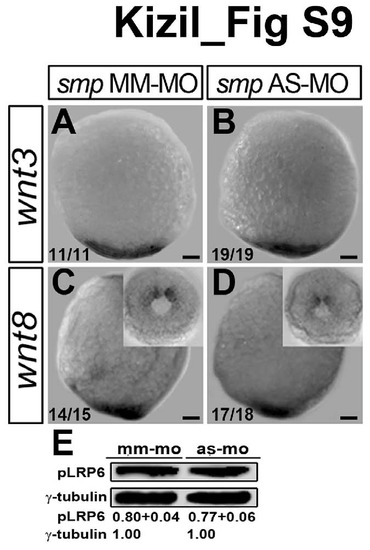
Smp knockdown does not affect ligand-induced activation of Wnt signaling, Related to Figure 2. (A) Expression of wnt3 in mismatch-control (MM-MO) embryos. (B) Expression of wnt3 in smp morphants (AS-MO). (C) Expressison of wnt8 in MM-control embryos. Inset shows posterior expression. (D) Expression of wnt8 in smp morphants. Inset shows posterior expression. (E) Western blots of phospho-Lrp6 from MM controls and smp morphants. Values indicate relative abundance. Numbers in the lower left corners in panels indicate the number of embryos with the depicted expression patterns to the total number of embryos. Scale bars equal 100 μm. |
|
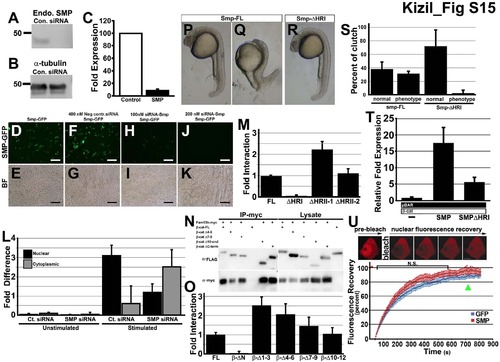
Reduction of SMP protein by transfection of Smp siRNA, Related to Figure 4. (A) Western blot of immnoprecipitation pull down of β-catenin with different mutants of the Smp protein. (B) Western blot of antibody to the endogenous SMP protein in control siRNA- and SMP siRNA-transfected HEK293T cells. (C) Same blot samples probed with α-tubulin antibody. (D) Measurement of fold expression detected from Western blot of siRNA knockdown experiment. (E-L) Expression of GFP fused to the C-terminal of Smp. (E) Smp-GFP alone and (F) Bright field image of (E). (G) Smp-GFP in cells transfected with control siRNA. (H) bright field of (G). (I) SMP-GFP fluorescence in the cells transfected with 100 nM siRNA to Smp. (K) SMP-GFP in cells transfected with 200 nM of siRNA to Smp. (M) Lucifierase reporter assay for control β-catenin, Smp-FL and Smp delta homology region (I). (N) Fluorescence recovery curves after photobleaching mCherry-β-catenin in nucleus of cell transfected either with GFP or Smp. Scale bars equal 100 μm. |