- Title
-
Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival
- Authors
- Zhang, D., Golubkov, V.S., Han, W., Correa, R.G., Zhou, Y., Lee, S., Strongin, A.Y., Dong, P.D.
- Source
- Full text @ Dev. Biol.
|
2F11 mAb labels hepatopancreas progenitors during organogenesis. (a) and (a′) Ventral view of a 20 hpf Tg(sox17:GFP) foregut showing broad low 2F11 labeling (turquoise arrowheads), and higher labeling in dorsal pancreas cells (turquoise arrows) and the pronephric ducts (white arrows). (b) and (c′) Lateral view of 26 hpf Tg(sox17:GFP) (b) and Tg(neuroD:EGFP) (c) foregut showing 2F11 labels the dorsal pancreas (dP) with a single focal plane shown in (c′′) and (c′′′). (d)-(g′′′) Ventral view of the Tg(sox17:GFP) foregut with Prox1 and 2F11 labeling at 30, 34, 42 and 48 hpf; (d′′′), (e′′′), (f′′′), (g′′′) digital sections along X, Y, and Z axes showing higher 2F11 staining on the ventral side of the endoderm (green lines indicate the positions of the X-axis sections, right insets; red lines indicate the positions of the Y-axis sections, top insets, a larger image of (f′′′) is shown in Fig. S1). (d)-(d′′) 2F11 staining increases in the Prox1+ budding liver (L) at 30 hpf. (e)-(e′′) At 34 hpf, 2F11 labeling increases in the Prox1+ ventral pancreas (vP) with broad cellular localization. Note the elevated staining in cells between the liver and ventral pancreas (white arrowhead). (f) and (f′′) At 42 hpf, 2F11 labeling increases further in the cells between liver and ventral pancreas with broad cellular localization. (g) and (g′′) At 48 hpf, 2F11 labels the entire hepatopancreas system with elevated staining and non-nuclear localization (white arrowheads) in cells between the liver and pancreas (P). (h) Ventral view of the Tg(ptf1a:GFP);Tg(lfabf:ds-Red) foregut at 80 hpf showing 2F11 labeling becomes restricted to the gallbladder (GB; yellow arrow) and ducts, including intrapancreatic ducts (white arrowheads), intrahepatic ducts (black arrowheads), and extra hepatopancreatic ducts (yellow arrowheads). (h′) Digital sections along X, Y, and Z axes showing higher 2F11 staining in the ventral region of the ducts. (i) and (i′) Higher magnification of (h′) (white square inset) showing 2F11 basal-lateral cellular localization in ductal columnar epithelium where it does not overlap with apically restricted Cadherin (yellow arrows). EXPRESSION / LABELING:
|
|
Anxa4 is bound by 2F11 and is expressed in tissues labeled by 2F11. (a) and (b) A 2F11 immuno-precipitated band (arrow in (a)) was analyzed by peptide mass fingerprinting, yielding peptide sequences (colored text in (b)) that match zebrafish Anxa4. (c)-(g′) Anxa4 is expressed in the developing zebrafish foregut (orange arrow), pronephric ducts (magenta arrowheads), dorsal pancreas (yellow arrows), liver bud (red arrowheads), lateral lines (turquoise arrowheads), and hatching gland (green arrowheads). (h)-(k′) The ventral view of Tg(sox17:GFP) with anxa4 transcript (h) and (j) or 2F11 labeling (i), (i′), (k) and (k′) of the liver (L), ventral pancreas (vP), and dorsal pancreas (dP) at 34 hpf (h) and (I′) and 47 hpf (j) and (k′). (l) and (m′) At 80 hpf, both anxa4 transcripts (l) and 2F11 (m) and (m′) are found in the gallbladder (GB), hepatopancreatic ducts, including extrahepatic duct (EHD), common bile duct (CBD), extrapancreatic duct (EPD), intrapancreatic duct (IPD) and intrahepatic duct (IHD), intestinal secretory cells (yellow arrowheads), and mesenchymal cells around the swim bladder (turquoise arrowheads). EXPRESSION / LABELING:
|
|
Knockdown of Anxa4 reduces 2F11 staining. (a) Diagram of anxa4 transcripts to depict targets of MOs and RT-PCR primers (green arrows). MO1 is targeted to block translation initiation in Exon 3 and MO2 is targeted to block the splice donor site at the Exon 4/Intron 4 boundary, predicted to cause skipping of Exon 4, leading translation from Exon 3 directly to Exon 5. (b) RT-PCR (with primers indicated in (a)) of control and MO2 morphant embryos showing the predicted 157 bp PCR product from the mis-spliced transcript (red arrow) and a reduction of the 245 bp PCR product from the normally-spliced transcripts (blue arrow) in morphants. (c) The sequence of the mis-spliced transcript confirms Exon 4 skipping and indicates a premature stop codon in Exon 5. (d) Both MOs lead to reduced 2F11 staining in a western blot of whole embryo lysate. (e)-(g′) Ventral views of 48 hpf Tg(sox17:GFP) showing that Anxa4 morphants have reduced 2F11 labeling in the entire hepatopancreas area compared to the control. L, liver; P, pancreas; dP, dorsal pancreas. EXPRESSION / LABELING:
|
|
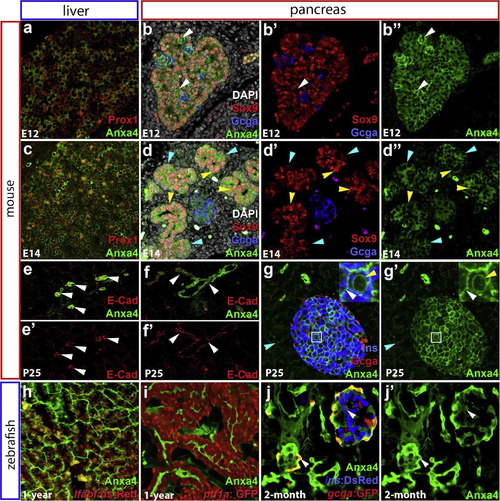
Conserved zebrafish and mouse Anxa4 expression in the liver and pancreas. (a)-(g′) Immunofluorescent labeling of Anxa4 in the liver and pancreas of E12, E14, and P25 mice. From E12 to E14, Anxa4 is detected in Prox1+ liver cells (a) and (c), and in all pancreatic cells (b) and (b′′); (d) and (d′′), including Sox9+ or Glucagon+ cells and cells negative for both Sox9 and Glucagon (white arrowheads in b-b′′). At E14, both the Sox9 high (yellow arrowheads) and low (blue arrowheads) expression cells are positive for Anxa4. In adult mice, Anxa4 is expressed the intrahepatic biliary ducts (e) and (e′); white arrowheads) and intrapancreatic ducts (f) and (f′); white arrowheads), co-localizing with ductal expression of E-Cadherin. Anxa4 is also expressed in pancreatic endocrine islets (g) and (g′), with localization to the membrane (white arrowhead) and nuclear envelope (yellow arrowhead). Low level Anxa4 is expressed in acinar cells (blue arrowheads) (h)-(j′) Immunofluorescent labeling of Anxa4 in the zebrafish liver (h) and pancreas (I) of one-year old Tg(ptf1a:GFP);Tg(lfabf:ds-Red), and pancreas of 2-month old Tg(ins:dsRed);Tg(gcga:GFP), showing Anxa4 is expressed in the networks of intrahepatic (h) and intrapancreatic (i) and (j′) ducts as well as pancreatic islets (j), (j′) single optical section, white arrowheads). EXPRESSION / LABELING:
|
|
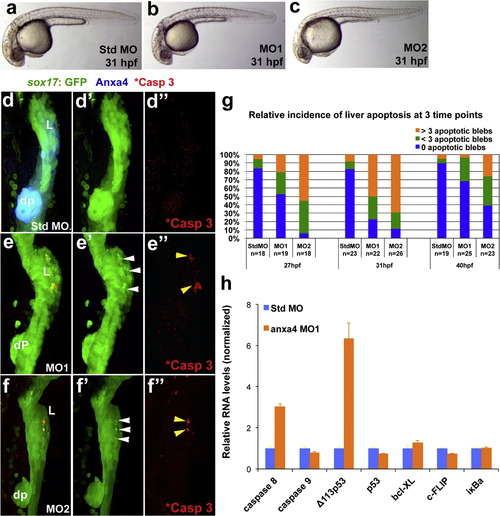
Anxa4 knockdown causes stage and tissue specific apoptosis in the developing foregut. (a)-(c) Bright field microscopy of 31 hpf standard control MO (a), Anxa4 MO1 (b) and Anxa4 MO2 (c) morphant zebrafish embryos showing a mild developmental delay in the Anxa4 morphants. (d)-(f′′) The ventral views of 31 hpf Tg(sox17:GFP) foregut stained for Anxa4 and cleaved Caspase 3 (*Casp 3). In Anxa4 morphants (MO1 in (e)-(e′′); MO2 in (f) and (f′′), but not in control morphants (d) and (d′′), Anxa4 labeling is mostly lost (e) and (f) and patches of GFP+ apoptotic blebs can be found restricted to the Anxa4 morphant liver buds (white arrowheads). A subset of these GFP blebs are also positive for cleaved Caspase 3 (yellow arrowheads). (g) Graph summarizing relative incidence of liver apoptosis in anxa4 morphants, which peaks at 31 hpf. (h) Graph of a representative set of real-time RT-PCR results showing that in 27 hpf Anxa4 MO1 morphants, there is elevated expression of caspase 8 and Δ113p53. |
|
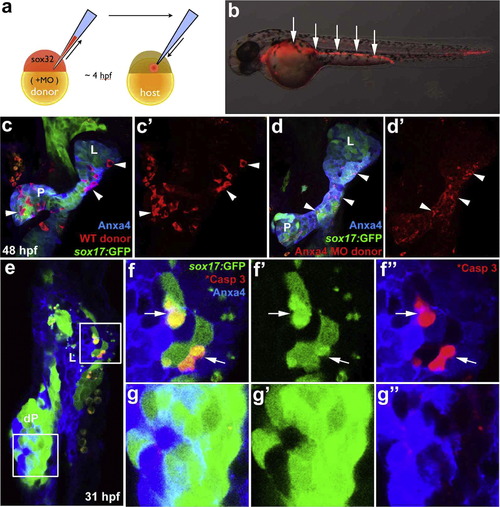
Anxa4 is cell-autonomously required for liver cell viability. (a) Diagram depicts endoderm transplantation strategy. Donor embryo is injected with sox32mRNA to drive endoderm fate, rhodamine dextran for labeling, and MOs to knock down gene expression. At 4 hpf, donor cells are transplanted into a host embryo, where they will contribute primarily to the host endoderm. (b) The mosaic 35 hpf host embryo obtained after the endoderm transplantation contains rhodamine dextran labeled donor endoderm cells (white arrows). (c) and (d′) The ventral view of 48 hpf Tg(sox17:GFP) foregut with rhodamine dextran labeled WT (white arrowheads in (c) and (c′) or anxa4 morphant donor cells (d) and (d′). L, liver; P, pancreas. Note that rhodamine labeling (white arrowheads in (d) and (d′)) appears scattered and not restricted within individual cells in the host embryo with Anxa4 morphant donor cells. (e)-(g′′) A single optical section (ventral view) of a 31 hpf mosaic host embryo with Anxa4 morphant cells from a Tg(sox17:GFP) donor embryo showing GFP blebs in the liver (upper inset and (f) and (f′′) but not dorsal pancreas (lower inset and (g) and (g′′). Subsets of GFP positive morphant donor cells are also positive for the cleaved Caspase 3 (white arrows) in the liver but not the dorsal pancreas. L, liver; dP, dorsal pancreas. |
|
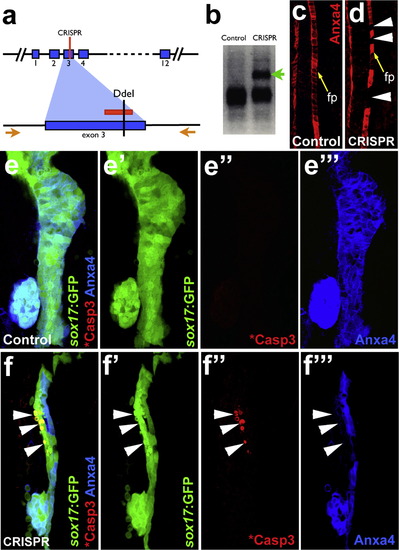
Mosaic disruption of theanxa4gene by CRISPR/Cas9 leads to apoptosis in the developing liver. (a) The CRISPR guide RNA is designed to target a region in Exon 3 of anxa4, which contains a DdeI restriction enzyme site. The red lines indicate the guide RNA target site. Orange arrows indicate the primers for the genotyping PCR. (b)-(d) Effective anxa4 disruption by CRISPR/Cas9 was confirmed by DdeI digestion of the PCR products (b) and by Anxa4 antibody labeling of the floor plate (c) and (d). An uncut band (green arrow in (b)) and the floor plate cells without Anxa4 expression (white arrowheads in (d)) are found only in injected embryos. Yellow arrows point to floor plate (fp) in (c) and (d). (e) and (f′′′) Confocal projections showing that apoptosis, indicated by GFP blebs and cleaved Caspase3 labeling, occurs in a subset of liver cells where Anxa4 expression is disrupted in the CRISPR injected embryos (white arrows) but not in control embryos. |
Reprinted from Developmental Biology, 395(1), Zhang, D., Golubkov, V.S., Han, W., Correa, R.G., Zhou, Y., Lee, S., Strongin, A.Y., Dong, P.D., Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival, 96-110, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.