- Title
-
Retinoic acid plays an evolutionarily conserved and biphasic role in pancreas development
- Authors
- Huang, W., Wang, G., Delaspre, F., Vitery, M.D., Beer, R.L., Parsons, M.J.
- Source
- Full text @ Dev. Biol.
|
Notch and RA signaling regulate the induction of 2° islet cells in a manner consistent with synergy. (A–D) Confocal z-stack projections of larval Tg(neuroD:GFP)nl1 pancreata dissected at 5 dpf. Larvae were treated with (A) DMSO, (B) RO4929097 (RO) 0.625 μM, (C) DEAB 25 μM and (D) DEAB 25 μM+RO 0.625 μM, from 3–5 dpf. 2° Islet cells (green) in the tail region of pancreata are indicated by arrowheads. Pancreata are outlined in white dashed lines. Scale Bar, 100 μm. (E) Average number of 2° islet cells per 5 dpf larval pancreas. N=number of larval pancreata quantified. Error Bar represents standard error around the mean (SE). Significance, **p<0.001. |
|
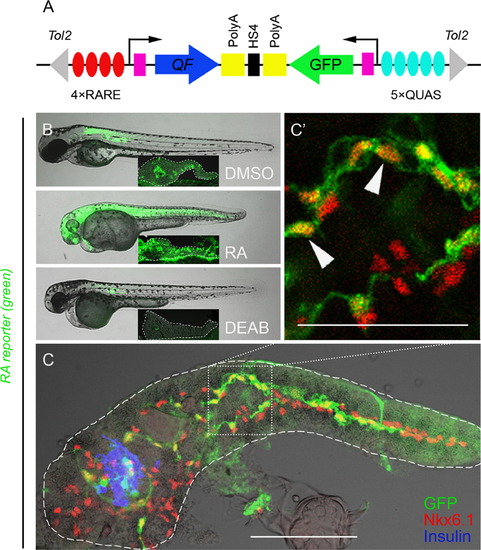
PNCs are RA-responsive. (A) Schematic of the RA reporter, 4xRARE-cFos:QF; QUAS:GFP. Expression of transcription factor QF (blue arrow) is driven by 4 RA-responsive elements (4xRARE, 4 red ovals). In the opposite orientation, GFP coding sequence (green arrow) is cloned downstream of 5 QF upstream activating sites (5xQUAS, 5 cyan ovals). QF binds to QUAS sequence to activate GFP expression. HS4 Insulator sequence (black rectangle) prevents the interference between the two transcriptional units. Pink rectangles represent cfos minimal promoters. The transgene is flanked by Tol2 arms (gray triangles). (B and C) Fluorescence images of RA reporter larvae. The channel of green fluorescence shows the RA reporter signal. (B) Compared to DMSO control, 3 dpf larvae treated with exogenous RA or DEAB increase or decrease their RA reporter signal, respectively. The larvae were incubated from 1–3 dpf with either 10 μM RA, 25 μM DEAB or DMSO. Insets show images of dissected pancreata from treated larvae at 5 dpf, outlined in white dashed lines. (C) RA-responsive cells are Nkx6.1+ in the larval pancreas. A single-plane confocal image of a larval pancreas dissected at 5 dpf and immunostained for Nkx6.1 and Insulin. Arrowheads indicate examples of GFP+ Nkx6.1+ cells. Scale bar, 100 μm. EXPRESSION / LABELING:
|
|
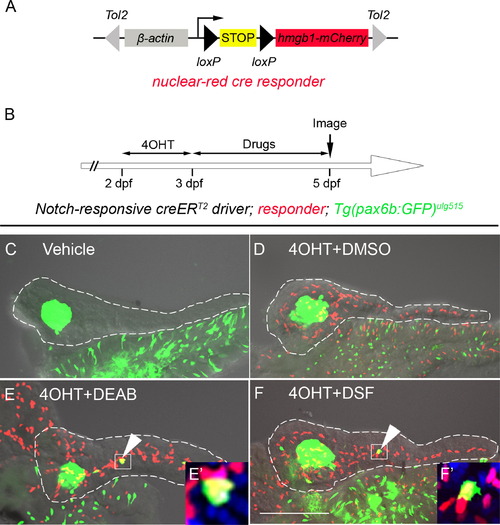
Inhibition of RA signaling induces PNCs to differentiate into endocrine cells. (A) Schematic of the nuclear red cre responder, Tg(β-actin:loxP-stop-loxP-hmgb1-mCherry)jh15. The β-actin promoter/enhancer drives ubiquitous expression. Active Cre induces recombination between the two loxP sites (black triangles) removing the STOP signal (yellow box) and allowing production of Hmgb1-mCherry, a nuclear-red fluorescent protein. The transgene is flanked by Tol2 arms (gray triangles). (B) Experimental timeline of 4OHT-dependent fate mapping. 4OHT was applied 2–3 dpf. Larvae were then exposed to drugs from 3–5 dpf. (C–F) Confocal z-stack projections of dissected pancreata from larvae transgenic for the Notch-responsive creERT2driver, Tg(Tp1glob:creERT2)jh12, the nuclear red cre responder and the endocrine marker, Tg(pax6b:eGFP)ulg515. These triple transgenic larvae were treated with vehicle (C), 4OHT+DMSO (D), 4OHT+DEAB (E), and 4OHT+DSF (F). The presence of lineage-traced cells (red nuclei) is dependent on 4OHT treatment (D–F). 2° islet cells can be detected in the tail region of larval pancreata after treatment with either DEAB (E) or DSF (F). (E′ and F′) The precocious 2° islet cells are positive for both GFP and mCherry, indicating their PNC origin. Nuclei are counterstained with DAPI (blue). White dashes outline pancreata. Scale Bar, 100 μm. |
|
Overexpression of dnRAR blocks pancreatic field specification. (A–D) The results of whole mount in situ hybridization to detect ins and trypsin expression in, dnRAR cre responder transgenic larvae at 72 hpf. Expression of ins and trypsin demonstrates normal pancreatic field specification. (A and B) Negative control larvae (CT). (C and D) Larvae previously injected with cre mRNA at the one-cell stage and displaying the failure of pancreas specification in presence of cre-dependent dnRAR-GFP. Arrows indicate the location of larval pancreata. EXPRESSION / LABELING:
|
|
PNC differentiation is regulated by endogenous RA signaling. (A and B) in situ hybridization for neuroD expression to detect 2° islets in 5 dpf pancreata from larvae transgenic for both the Notch-responsive creERT2driver, Tg(Tp1glob:creERT2)jh12, and the dnRAR cre responder, Tg(ubb:loxP-eCFP-loxP-dnRAR-GFP)jh39. These larvae had been incubated at 2 dpf with either vehicle (A) or 4OHT (B). (C) Average number of 2° islets/pancreas in vehicle or 4OHT treated larvae calculated by counting neuroD expressing islets. (D–G) Fluorescence immunostaining to detect hormone-expressing cells (insulin+/glucagons+/somatostatin+), facilitating quantification of 2° islet cells in larvae that were treated from 2–5 dpf with either vehicle (D), or 4OHT (E), or vehicle+DEAB (F). (E′′) enlarged image of (E′). (G) Average number of 2° islet cells/pancreas in vehicle, 4OHT, and vehicle+DEAB treated larvae calculated by counting for the number of hormone-expressing cells. **p<0.001. White dashes outline pancreata. 2° Islets in the tail region of the pancreas are indicated by arrowheads. N=number of larval pancreata quantified. Error Bar=SE. Scale Bar, 100 μm. |
|
Exogenous RA prevents the normal formation of secondary islets in larval fish. (A–B) Confocal z-stack projections of larval Tg(neuroD:GFP)nl1 pancreata dissected at 8 dpf. Larvae were treated with DMSO and different concentrations of RA from 5 to 8 dpf. 2° islet cells (green) in the tail region of pancreata are indicated by arrowheads. Pancreata are outlined in white dashed lines. Scale Bar, 100 μm. (C) Average number of 2° islet cells per 8 dpf larval pancreas. N=number of larval pancreata quantified. Error Bar represents standard error around the mean (SE). Significance, ***p<0.001. |
|
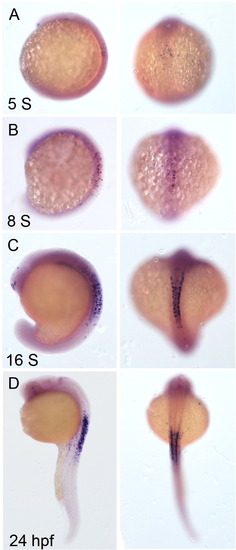
Characterization of the new RA reporter. (A–D) Expression of gfp mRNA was detected by whole mount in situ hybridization. Shown are images of the embryos of RA reporter at different stages: (A) 5 Somites (S), (B) 8S, (C) 16S, and (D) 24 hpf. Left panels, lateral views of the embryos. Right panels, dorsal views. |
|
Nkx6.1 marks the PNCs in the larvae. (A–C) Projections of confocal z-stacks of larval Notch reporter (nuclear red) at 5 dpf. The larval pancreata were dissected out after being fixed with 4% PFA, and immunostained for Nkx6.1. (A) A merged image of the channels for Nkx6.1 (green) and mCherry (red). Nuclear-localized Nkx6.1 signal mainly appears in the intrapancreatic duct region, which is the location of the mCherry-labeled PNCs. Most of the Nkx6.1+ cells are also positive for mCherry. (B) The channel for Nkx6.1 staining. (C) The channel for Notch reporter (nuclear-red). |
|
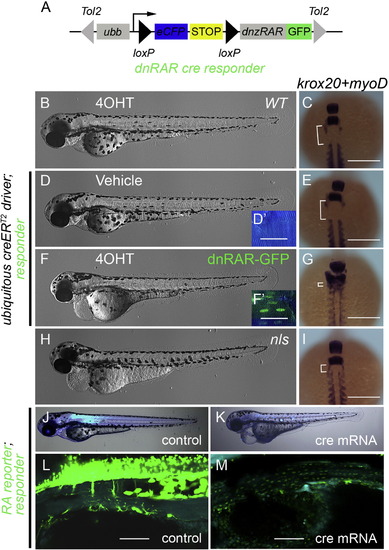
Over-expression of dominant-negative zebrafish RA receptor α inhibits endogenous RA signaling. (A) Schematic of the dnRAR cre responder, Tg(ubb:loxP-eCFP-loxP-dnRAR-GFP)jh39. Tol2 arms (gray triangles) flank the whole expression cassette. Ubb promoter/enhancer drives ubiquitous expression. Active cre induces recombination between the two loxP sites (black triangles), removing the STOP signal (yellow box), and allowing production of the dnRAR-GFP fusion protein. (B, D, F, and H) Shows phase contrast images of whole 3 dpf larvae. (D′ and F′) Fluorescent images of 3 dpf larval trunk. Before recombination cells are labeled with eCFP (blue) and after cre activity with nuclear GFP (green). Scale Bar, 25 μm. (C, E, G, and I) Dorsal views of 11-somite stage embryos following whole mount in situ hybridization to detect krox20 and myoD expression. Scale Bar 200 μm. Different experimental conditions as follows: (B and C) wild-type fish incubated from 4–72 hpf with 4OHT. (D–G) Fish transgenic for the ubiquitous creERT2driver and the dnRAR cre responder, were treated with vehicle (D and E) or 4OHT (F and G) from 4–72 hpf. For phenotypic comparison to larvae lacking ALDH1a2 activity, ALDH1a2nls/nls mutant larvae were analyzed (H and I). After 4OHT treatment, unlike controls (B and D), larvae transgenic for the ubiquitous creERT2driver and dnRAR cre responder have nuclear localized GFP, due to the expression of dnRAR-GFP (F′), and a gross morphological phenotype (F) similar to the nls mutant (H). The deleterious phenotype associated with compromised RA signaling includes patterning defects (G and I) when compared to controls (C and E). These defects can be visualized by a decrease in distance (white bracket) between the most anterior somite (marked by myoD expression) and rhombomere 5 (marked by krox20 expression) (Oxtoby and Jowett, 1993; Weinberg et al., 1996). (J–M) Fluorescent images of the larvae of dnRAR cre responder; RA reporter at 72 hpf. Cre mRNA was injected at one-cell stage to induce widespread expression of dnRAR-GFP fusion protein (K and M), while non-injected larvae were used as controls (J and L). Cytoplasmic-GFP signal (clearly seen in L) indicates the activity of RA reporter, and nuclear-GFP signal (visible in M) indicates the induction of dnRAR-GFP fusion protein. Induction of dnRAR-GFP greatly diminishes activity of the RA-reporter. Scale bar, 50 μm in L and M. |
|
Injection of cre mRNA induces expression of dnRAR in the larvae of dnRAR cre responder, but does not change the expression pattern of ins and trypsin in the larvae of nuclear-red cre responder. (A and B) Fluorescent images of the larval somites of dnRAR cre responder at 72 hpf. (A) Non-cre mRNA injected larvae (negative control). (B) cre mRNA injected larvae. cre mRNA was injected at one cell stage. Widespread expression of nuclear-GFP can be detected in the larvae with cre mRNA injection, but not in the control. (C and D) Fluorescent images of the larvae somites of nuclear-red cre responder at 72 hpf. (C) Non-cre mRNA injected larvae. (D) cre mRNA injected larvae. cre mRNA injection robustly induces the recombination of LoxP sites, indicted by the widespread expression of nuclear-mCherry, but not in the non-injected controls. Scale bar in A–D, 10 μm. (E and F) In situ hybridization images of the nuclear-red cre responder larvae at 72 hpf. Cre mRNA was injected into the embryos at one-cell stage. (E) ins expression in the larvae. (F) trypsin expression in the larvae. Injection of cre and subsequent recombination does not affect pancreatic development. |
Reprinted from Developmental Biology, 394(1), Huang, W., Wang, G., Delaspre, F., Vitery, M.D., Beer, R.L., Parsons, M.J., Retinoic acid plays an evolutionarily conserved and biphasic role in pancreas development, 83-93, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.