- Title
-
Distinct tissue-specific requirements for the zebrafish tbx5 genes during heart, retina and pectoral fin development
- Authors
- Pi-Roig, A., Martin-Blanco, E., Minguillón, C.
- Source
- Full text @ Open Biol.
|
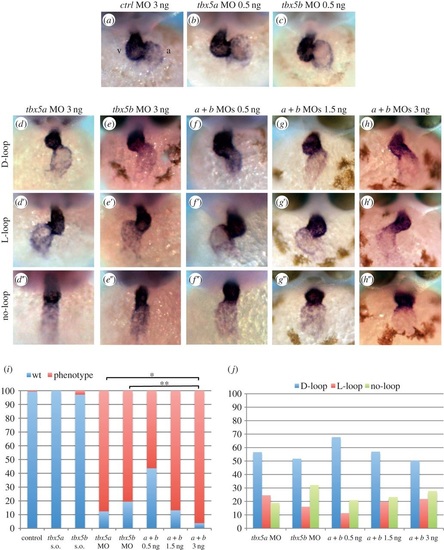
Knock-down of tbx5 genes causes cardiac looping defects. (a-c) Embryos injected with control MO or sub-optimal concentrations of tbx5a or tbx5b MOs. (d-d′′) tbx5a morphant phenotypes. (e-e′) tbx5b-morphant phenotypes. (f-h′′) Double knock-down of tbx5 genes (0.5 ng each MO (f-f′′), 1.5 ng each MO (g-g′′) and 3 ng each MO (h-h′′)). (i) Quantification of the degree of looping phenotypes: wt, complete; phenotype, incomplete looping s.o., sub-otimal. (j) Quantification of the looping orientation phenotypes. A χ2 statistic has been calculated to assess significant differences between groups (**p < 0.001, *p < 0.05). Images are frontal views of 48 hpf embryos, and myl7 expression is used to highlight the developing heart. PHENOTYPE:
|
|
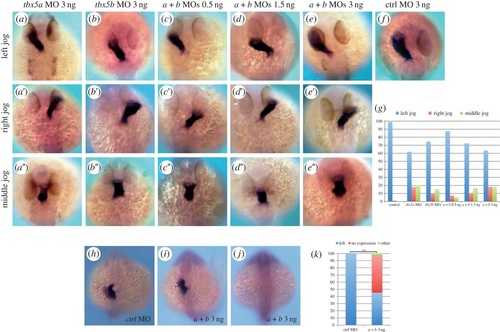
tbx5 morphants exhibit cardiac jogging and lefty2 expression defects. (a-a′′) tbx5a morphant phenotypes. (b-b′′) tbx5b morphants. (c-e′′) Left, right and middle jog phenotypes obtained by co-injection of tbx5a and tbx5b MOs at 0.5 ng (c-c′′), 1.5 ng (d-d′′) or 3 ng (e-e′′) of each MO. (f) Control (ctrl) morphant. (g) Quantification of the phenotypes. (h-j) lefty2 expression in control (h) and double-morphant (i,j) 22-somite stage embryos. (k) Quantification of the phenotypes. A χ2 test has been used to assess significant differences between groups (**p < 0.001). All images are dorsal views with anterior to the top, and myl7 expression is used to highlight the developing heart tube in a-f. EXPRESSION / LABELING:
PHENOTYPE:
|
|
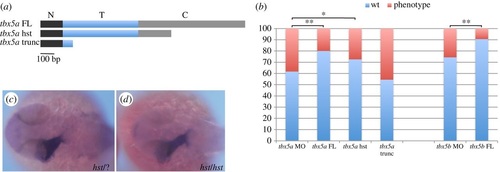
The hst mutation is a hypomorphic allele with regards to cardiac laterality. (a) tbx5a variants generated to perform the rescue experiments: a tbx5a full-length (tbx5a FL) version includes the whole N-terminal (N, black rectangle), T- (T, blue rectangle) and C-terminal (C, grey rectangle) domains; a heartstrings version (tbx5a hst) containing the whole N-terminal domain and T-domain and a truncated C-terminal domain; and a tbx5a severely truncated version (tbx5a trunc) that contains the whole N-terminal domain and a truncated T-domain. (b) Quantification of the rescue experiments (wt, left jog; phenotype, right and middle jog). A χ2 statistic has been calculated to assess significant differences between groups (**p < 0.01, *p < 0.05). (c,d) Dorsal views with anterior towards the left of 26 hpf +/+ or +/hst (c) and hst mutant (d) embryos showing normal leftward jogging of the embryonic heart tube highlighted by myl7 expression. PHENOTYPE:
|
|
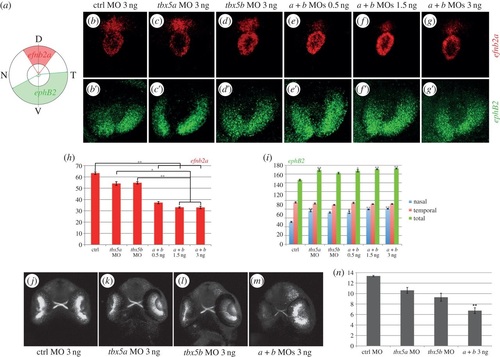
tbx5 genes are required for dorsoventral retina organization. (a) Schematics of our quantification method. Expression of efn2a and ephB2 in control (ctrl) embryos (b-b′), tbx5a morphants (c-c′) and tbx5b morphants (d-d′). (e-g′) Expression of efn2a and ephB2 in embryos co-injected with different concentrations of both tbx5a and tbx5b MOs. (h-i) Quantification of the results obtained for the expression of efn2a (h) and ephB2 (i). (j-m) Retinal projections of 48 hpf ath5:GFP embryos injected with control, tbx5a, tbx5b or tbx5a and tbx5b MO. (n) Optic nerve diameter quantifications. Data are represented as the mean ± s.e. A Kruskal-Wallis test was used to determine statistical differences among experimental groups (*p < 0.05, **p < 0.001). D, dorsal; N, nasal; T, temporal; V, ventral. |
|
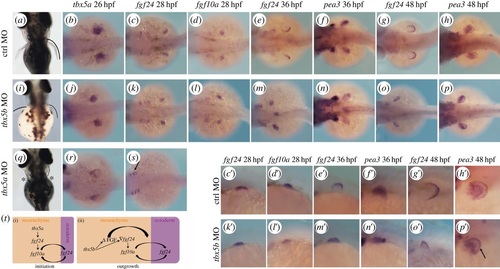
tbx5b knock-down causes a delay in pectoral fin growth. (a,i,q) Pectoral fin morphology at 3 dpf. Dorsal views are shown with anterior to the top. (b-h) Expression of the developing pectoral fin markers in control (ctrl) MO-injected embryos. (c′-h′) Higher magnifications of (c-h). (j-p) Pectoral fin markers expression in tbx5b-morphant embryos. (k′-p′) Higher magnifications of (k-p). (r,s) tbx5a morphants. (t) Model for the differential requirements for the tbx5 genes during pectoral fin development. b-h, j-p,r,s are dorsal views with anterior to the left. c′-h′,k′-p′ are lateral views with anterior to the left. EXPRESSION / LABELING:
PHENOTYPE:
|