- Title
-
Complexes of Usher proteins preassemble at the endoplasmic reticulum and are required for trafficking and ER homeostasis
- Authors
- Blanco-Sánchez, B., Clément, A., Fierro, J., Washbourne, P., Westerfield, M.
- Source
- Full text @ Dis. Model. Mech.
|
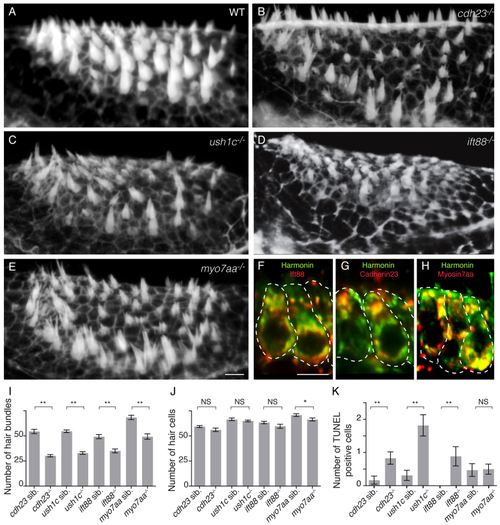
Zebrafish cdh23, ush1c, myo7aa and ift88 mutants have mechanoreceptor structural defects. (A–E) Confocal projections of anterior maculae labeled with phalloidin in (A) wild-type (WT) sibling, (B) cdh23tj264a, (C) ush1cfh293, (D) ift88tz288b and (E) myo7aaty220d mutants. (F–H) Confocal optical sections of double immunolabeling of (F) Harmonin (green) and Ift88 (red), (G) Harmonin (green) and Cdh23 (red), and (H) Harmonin (green) and Myo7aa (red). Individual hair cells are shown. (I) Quantification of total number of hair bundles. ne6 analyzed anterior maculae. (J) Quantification of total number of hair cells. ne5 analyzed anterior maculae. (K) Quantification of hair cell death in mutants and wild-type siblings. Number: average number of TUNEL-positive hair cells per macula. ne16 analyzed anterior maculae. Student’s t-test: **P<0.01, *P<0.05. NS, non significant. Black lines represent ± s.e.m. Scale bars: 5 µm. |
|
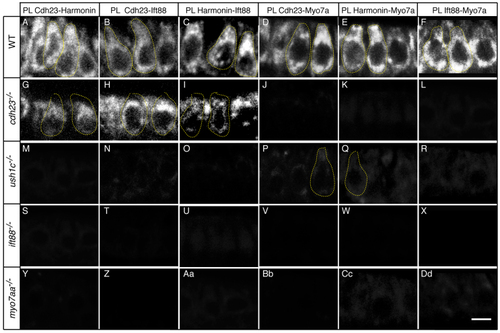
Cdh23 and Harmonin require Ift88 and Myo7aa for normal localization in hair cells. Immunolabeling in (A-D) wild-type siblings or (E-H) cdh23, (I-L) ush1c, (M-P) ift88 and (Q-T) myo7aa mutants of (A,E,I,M,Q) Cdh23, (B,F,J,N,R) Harmonin, (C,G,K,O,S) Ift88 and (D,H,L,P,T) Myo7aa. Confocal views of anterior macula hair cells are shown. Confocal sections are 0.8 µm thick, whereas the neck and basal region of the hair cell have an estimated diameter of 2-2.5 µm and 5-6 µm, respectively. Thus, not all confocal sections contain a hair bundle, and the observed shape of the hair bundle varies according to the mounting angle (see also supplementary material Fig. S3). Some individual hair cells are outlined by dotted lines. WT, wild type. Scale bar: 5 µm. EXPRESSION / LABELING:
|
|
Cdh23, Harmonin, Ift88 and Myo7aa proteins are in close proximity. Proximity labeling (PL) of (A,G,M,S,Y) Cdh23 and Harmonin, (B,H,N,T,Z) Cdh23 and Ift88, (C,I,O,U,Aa) Harmonin and Ift88, (D,J,P,V,Bb) Cdh23 and Myo7a, (E,K,Q,W,Cc) Harmonin and Myo7a, and (F,L,R,X,Dd) Ift88 and Myo7. (A–F) Wild-type (WT) siblings; (G–L) cdh23, (M–R) ush1c, (S–X) ift88 and (Y-Dd) myo7aa mutants. Confocal views of anterior macula hair cells. Some individual hair cells are outlined by dotted lines. Scale bar: 5 µm. |
|
Harmonin and Cdh23 bind directly to each other, but not to Ift88. (A) Co-immunoprecipitation. MDCK cells were transfected as indicated by (+) with combinations of DNA vectors encoding Harmonin-HA, mbn-Cdh23-GFP, Cdh23-GFP or IFT88-HA. Input lanes: 1,4,7,10,13. Unbound fraction lanes: 2,5,8,11,14. Bound fraction lanes: 3,6,9,12,15. Immunoprecipitation was performed with an antibody against Harmonin (α-Harm) or GFP (α-GFP). (B,C) Confocal sections of transfected MDCK cells. (B) Double immunolabeling of mbn-Cdh23-GFP (green) and Harmonin (red). (C) Double immunolabeling of mbn-Cdh23-GFP (green) and Ift88 (red). Harmonin-HA: full-length zebrafish Harmonin isoform A fused to HA-tag at the C-terminal. mbn-Cdh23-GFP: zebrafish Cdh23 membrane bound cytoplasmic domain fused to GFP at the C-terminal. Cdh23-GFP: zebrafish Cdh23 cytoplasmic domain fused to GFP at the C-terminal. Ift88-HA: full-length zebrafish Ift88 fused to HA-tag at the C-terminal. Scale bar: 7.5 µm. |
|
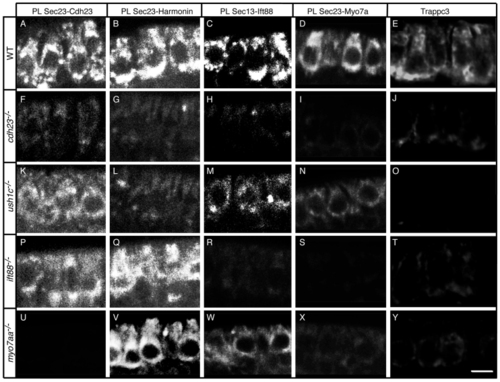
The Cdh23, Harmonin, Ift88 and Myo7aa protein complex is present at the ER and associated vesicles. Proximity labeling (PL) of (A–D) wild-type (WT) siblings, and (F–I) cdh23, (K–N) ush1c, (P–S) ift88 and (U–X) myo7aa mutants. Proximity labeling of (A,F,K,P,U) Sec23 and Cdh23, (B,G,L,Q,V) Sec23 and Harmonin, (C,H,M,R,W) Sec13 and Ift88, and (D,I,N,S,X) Sec23 and Myo7a (recognizes Myo7aa and possibly Myo7ab; see Materials and Methods). (E,J,O,T,Y) Immunolabeling of Trappc3 in (E) WT, and (J) cdh23, (O) ush1c, (T) ift88 and (Y) myo7aa mutants. Confocal views of anterior macula hair cells. Scale bar: A–D,F,I,K–N,P–S,U–X: 7.5 µm; E,J,O,T,Y: 5 µm. |
|
The Cdh23, Harmonin and Myo7aa protein complex is required for proper trafficking of USH2 proteins and can trigger ER stress when defective. (A–O) Immunolabeling of USH2 proteins in (A–C) wild-type (WT) siblings, and (D–F) cdh23, (G–I) ush1c, (J–L) ift88 and (M–O) myo7aa mutants with antibodies against Ush2a (A,D,G,J,M), Gpr98 (B,E,H,K,N) and Whirlinb (C,F,I,L,O). (P) Levels of hspa5 expression based on in situ hybridization technique in whole-mount larvae. For ush1c siblings n=57; ush1c/ n=51; cdh23 siblings n=35; for cdh23/ n=33; myo7aa siblings n=24; for myo7aa/ n=24 analyzed anterior maculae. (Q) Quantification of percentage area per hair cell containing KDEL expression. For ush1c siblings n=85; ush1c/ n=125; cdh23 siblings n=66; for cdh23/ n=60; myo7aa siblings n=60; myo7aa/ n=54 analyzed hair cells for each genotype. (R) Knockdown of cdk5 rescues ER-stress-induced cell death in the ush1c mutant. Bars show average number of TUNEL-positive cells. For ush1c siblings n=52; ush1c siblings + 4 ng cdk5-MO n=18; ush1c/ n=70; ush1c/ + 4 ng cdk5-MO n=70 analyzed anterior maculae. Average ± s.d. Statistics were conducted with Student’s t-test. **P<0.01. NS, non significant. Some individual hair cells are outlined by dotted lines. MO, morpholino. Scale bar: 5 µm. EXPRESSION / LABELING:
|
|
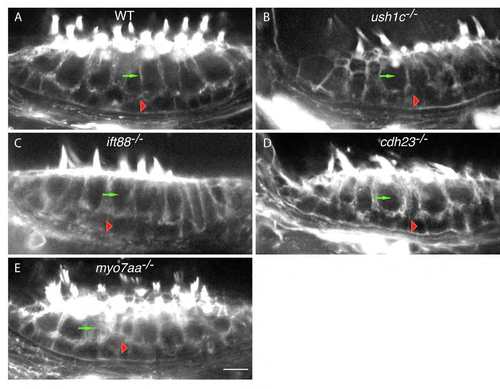
Epithelial organization of the anterior macula is unaffected in USH1 and Ift88 mutants. Phalloidin labeling of anterior maculae in (A) wild-type, (B) ush1c, (C) ift88, (D) cdh23, and (E) myo7aa animals. Green arrows indicate hair cells; red arrowheads indicate supporting cells. WT: wild-type. Scale bar: 5 µm. |
|
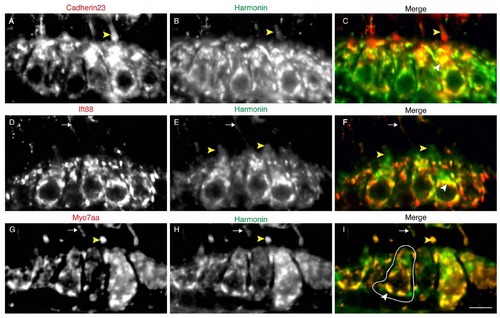
Colocalization of USH1 and Ift88 proteins in the hair cell. Double immunolabeling of Cdh23 (A), Ift88 (D), Myo7aa (G), and Harmonin (B,E,H). Panels C,F, I show merged panels of A and B, D and E, G and H, respectively. Fig. 1F,G are insets of panels C and F. White arrowheads: cell body colocalization. Yellow Arrowheads: hair bundle localization. Arrows indicate kinociliary labeling. A cell body is highlighted in white. Scale bar: 5µm. |
|
Cdh23, Harmonin, Myo7aa and Ift88 localize at the level of the mechanoreceptor. (A-I′) Wild-type zebrafish anterior maculae immunolabeled for Cdh23 (A,B), Harmonin (C,D,I), or Ift88 (E,F). As a control for the binding specificity of the AB Complex, primary antibody was omitted during the reaction (H). As a control for the specificity of the second antibody detection reaction, the first reaction was developed (I), then the AB complex was quenched and blocked (see material and methods), followed by omission of the other primary antibody during the reaction (I′). Both controls resulted in an absence of signal. Confocal sections focused on the hair bundle region. Note the appearance of the hair bundle depends on its spatial distribution along the anterior macula and on the mounting angle during image acquisition (compare panels A-G,I). Yellow arrowheads: hair bundle localization. Arrows: kinociliary localization. Cell bodies in different plans are highlighted in different colors. Scale bar: 5 µm. |
|
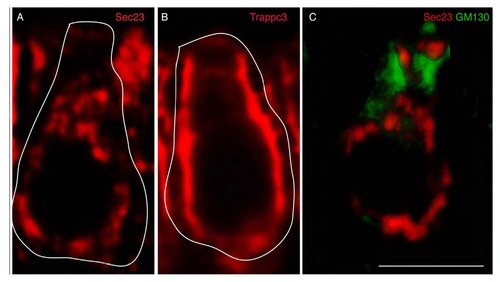
AB Complex detection method resolves cytoplasmic protein pools. Proteins that participate in the secretory pathway leave the ER via budding vesicles labeled by Sec23, a component of the COPII machinery (A). Trappc3 mediates fusion of Sec23 vesicles that form the ER-Golgi intermediate compartment (ERGIC) (B). After fusion, Sec23 and other COPII components return to the ER. Vesicles budded from the ERGIC then fuse with the membranes of the Golgi apparatus, labeled by GM130 (C). GM130 and Sec23 do not co-localize despite both having broad cytoplasmic labeling (C). This indicates that colocalization signals detected with the AB complex method are specific and not due to cross-reactivity or generalized labeling of the cell body. Some cell bodies are highlighted in white. Scale bar: 5µm. |
|
Cdh23 can reach the stereocilia in myo7aa mutants. (A,C) Specificity test for chicken anti-Cdh23 in wild-type sibling (A) and cdh23tj264a zebrafish mutant (C) using the AB complex detection method. A reminiscent weak signal is observed in the cdh23 mutant. This indicates that the tj264a mutation does not affect all cadherin 23 isoforms. (B,C) Cdh23 can be detected in the stereocilia of wild-type sibling (B) and myo7aa mutant (C) using a direct detection method.. In the direct detection conditions, quantification of the protein amount cannot be done due to saturation of the photodetector or the overall structure of the mechanoreceptor coupled to the lack of cytoplasmic signal in control experimental conditions. Arrowhead: hair bundle protein localization. Cell bodies are highlighted in different colors. Scale bar: 5µm. |
|
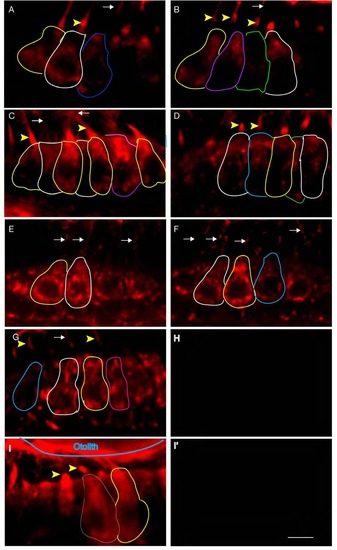
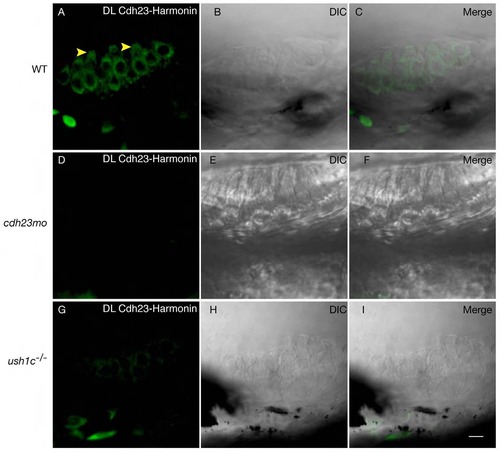
Proximity labeling of protein pairs is specific. Proximity labeling of Harmonin and Cdh23. (A) In wild-type anterior macula, Harmonin and Cdh23 are in close proximity in hair cell bodies. Note the apical localization of the signal (yellow arrowheads). (B,C): DIC channel and merge of panels A and B. (D-I) Negative controls. No significant signal was obtained when Cdh23 expression was knocked down by injection of cdh23 morpholinos (D) or in ush1c mutants (G). (E,H) DIC channel. (F,I) Merge of D and E, and H and I respectively. WT: wild-type; MO: morpholino. Scale bar: 5 µm. |
|
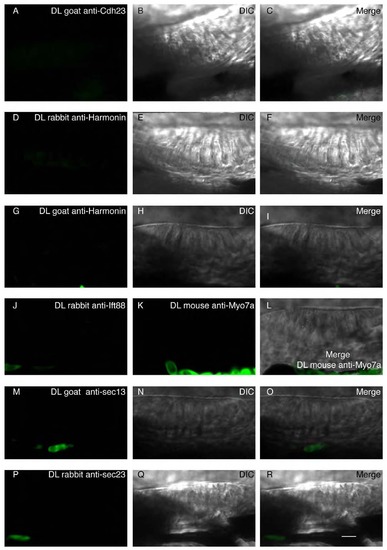
Proximity ligation control: Assay of nonspecific binding between proximity ligation probes in the absence of one of the primary antibodies during the reaction in WT conditions. (A,D,G,J,K,M,P) The proximity ligation reaction was performed with only one primary antibody: Goat anti-Cdh23 (A), rabbit anti-Harmonin (D), goat anti-Harmonin (G), rabbit anti-Ift88 (J), mouse anti-Myo7a (K), goat anti-Sec13 (M), or rabbit anti-Sec23 (P). (B,E,H,N,Q) single DIC channel showing a lateral view of the anterior macula. (C,F,I,L,O,R) Merged panels. Nonspecific signal was detected when only one primary antibody was added to the reaction. Scale bar: 5µm. |
|
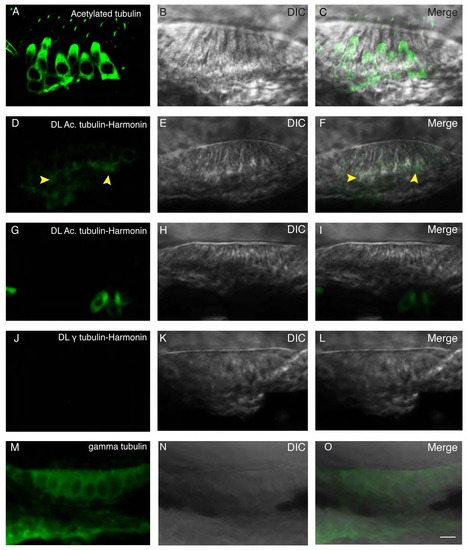
Proximity ligation assay indicates close physical proximity at the subcellular level. (A) Immunolabeling of acetylated tubulin. Acetylated tubulin fills up the cytoplasm and labels the kinocilium. (B) DIC channel. (C) Merge of panels A and B. (D) Proximity ligation assay of acetylated tubulin and Harmonin results in a restricted signal localized in the basal region of the hair cell, as indicated by the yellow arrowheads. (E) DIC channel. (F) Merge of panels D and E. (G) Proximity ligation assay of acetylated tubulin and Harmonin in ush1c mutant results in the absence of signal. (H) DIC channel. (I) merge of panels G and H. (J) Proximity ligation reaction of gamma tubulin and Harmonin resulted in no signal. (K) DIC channel. (L) Merge of panels J and K. (M) Immunolabeling of gamma tubulin showing a nonspecific signal filling the cytoplasm. (N) DIC channel. O: merge of panels M and N. Scale bar: 5µm. |
|
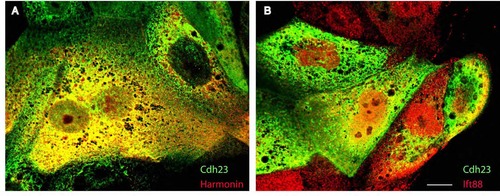
Subcellular colocalization of Harmonin, Cadherin23 and Ift88 in COS cells. (A) Immunolabeling of mbn- Cdh23-GFP (green) and Harmonin-HA (red). (B) Immunolabeling of mbn-Cdh23-GFP (green) and Ift88-HA (red). Scale bar: 5 µm. |
|
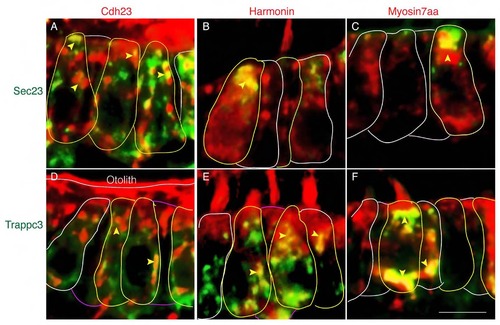
USH1 proteins colocalize with Sec23 and Trappc3. (A-C) Colocalization of Sec23 and Cdh23 (A), Sec23 and Harmonin (B), Sec23 and Myo7aa (C) in wild-type sibling. (D-F) Colocalization of Trappc3 and Cdh23. (D), Trappc3 and Harmonin (E), Trappc3 and Myo7aa (F) in wild-type sibling. Cell bodies are outlined. Yellow arrowheads indicate colocalization signal in the cell bodies. Scale bar: 5 µm. |
|
Loss of ERGIC in ush1 and ift88 mutants. (A-D) Wild-type siblings (WT). (E) cdh23 mutant. (F) ush1c mutant. (G) myo7aa mutant. (H) ift88 mutant. Loss of Trappc3 immunolabeling indicates a severe defect during the secretory process in ush1 and ift88 mutants. Scale bar: 5 µm. |
|
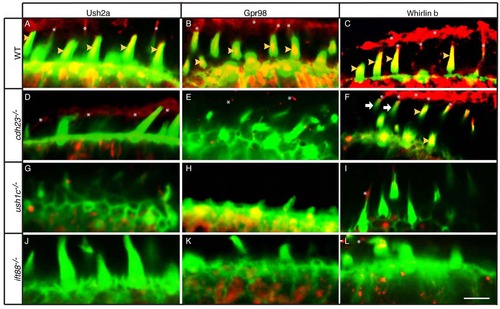
Hair bundle localization of USH2 Proteins is compromised in cdh23, ush1c, and ift88 mutants. (A-L) Phalloidin labeling (green) and immunolabeling (red) of USH2 proteins in (A-C) wild-type siblings or (D-F) cdh23, (G-I) ush1c, or (J-L) ift88 mutants with antibodies against Ush2a (A,D,G,J), Gpr98 (B,E,H,K), or Whirlinb (C,F,I,L). All images are single confocal sections at the level of the mechanoreceptor. Yellow arrowheads indicate stereociliary localization of USH2 protein. Asterisks indicate kinociliary localization of USH2 protein. White arrows (F) indicate hair bundles lacking Whirlinb in the cdh23 mutant. Scale bar: 5 µm. |
|
USH2 proteins accumulate in the ER in ush1c mutants. (A-D) Wild-type siblings. (E,F) ush1c mutants. (A,E) KDEL, a marker of the ER, is expanded in ush1c mutants. (B-D,F-H) USH2 proteins are stuck at the level of the ER in ush1c mutants. (B,F) Colocalization of Ush2a and KDEL. (C,G) Colocalization of Gpr98 and KDEL. (D,H) Colocalization Whirlinb and KDEL. USH2 proteins can be detected at low levels in the region of the ER (B-D). Colocalization signal between USH2 proteins and KDEL is remarkably increased in ush1c mutants. Some cell bodies are outlined. Scale bar: 5µm |
|
Mouse anti-Myo7a cross-reacts with zebrafish Myo7aa. (A) Antibody labeling in wild-type zebrafish anterior macula hair cells. (B) Labeling was greatly reduced in myo7aa mutants. WT: wild-type. Scale bar: 5 µm. |