- Title
-
Glycogen synthase kinase 3 inhibitors induce the canonical WNT/β-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma
- Authors
- Chen, E.Y., Deran, M.T., Ignatius, M.S., Grandinetti, K.B., Clagg, R., McCarthy, K.M., Lobbardi, R.M., Brockmann, J., Keller, C., Wu, X., Langenau, D.M.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
A secondary screen identifies lead compounds that suppress ERMS tumor growth in live zebrafish. (A) Schematic of the secondary screen completed in zebrafish transplanted with fluorescent-labeled ERMS. (B–G) Pretreatment images for DMSO (B), sunitinib (C), and BIO (D), with corresponding posttreatment images (E, F, and G, respectively). Tumor volume is indicated by the heat map (Right). (Scale bar, 2 mm.) (H) Summary of tumor volume changes in animals treated with compounds that inhibit RAS-signaling in embryonic zebrafish (blue), representative compounds of major classes of hits identified in the human differentiation screen (yellow), or common hits from both screens (green). Error bars equal SD. *Statistical significance by Student t test, with P < 0.05. |
|
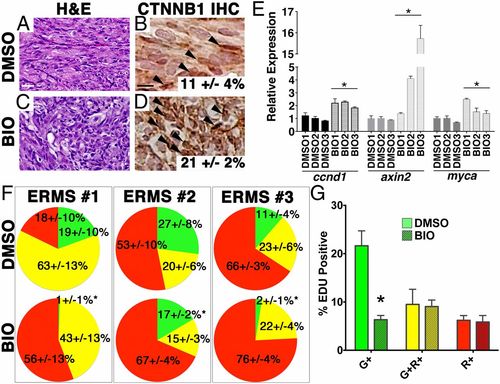
BIO activates canonical WNT/β-catenin signaling and induces differentiation of ERMS cells in vivo. (A–D) H&E-stained sections (A and C) and immunohistochemistry for β-catenin (B and D) of ERMS-bearing fish after 7 d of drug treatment with BIO (300 nM) or DMSO (vehicle control). (Scale bar, 25 μM.) (E) Quantitative RT-PCR analysis. (F) Pie charts showing the relative percentage of myf5:GFP+/mylz2:mCherry TPCs (green), myf5:GFP+/mylz2:mCherry+ (yellow), and late-differentiating myf5:GFP/mylz2:mCherry+ cells (red) for three independent ERMS as assessed by FACS (n > 3 animals per experimental arm and ±SD). *Statistically significant differences between DMSO and BIO treatments (Student t test, P < 0.05). (G) Summary of EDU staining of DMSO- or BIO-treated tumors. Each error bar indicates SEM of three tumors from each treatment group. *P < 0.005, Student t test. |
|
Lead compounds functionally alter cancer-specific processes. (A) RD cells were treated with lead compounds from the zebrafish secondary screen at four concentrations in the range of IC50. Cell growth/viability were assessed by cell titer glo after 4 d of drug treatment. Relative growth was normalized to cell numbers contained within each well at the beginning of the experiment. *Statistical significance with P < 0.05. (B) Lead compounds that affected growth (rapamycin, BIO, and trichostatin A) were further tested for their role in regulating apoptosis using a caspase glo assay. Average OD values from triplicate plating in an assay are shown. Each error bar denotes SD. *P value < 0.05, Student t test comparing test compound and DMSO. (C) Summary of angiogenesis assay. Tg(fli1:GFP embryos) were treated with DMSO and lead compounds (50 nM–10 μM) starting at early bud stage for 48 h. The effect of each compound on angiogenesis of intersegmental and tail vessels was assessed. Red indicates compounds that inhibit angiogenesis in fli1:GFP embryos. (D–F) Representative images of DMSO-treated (D), cediranib-treated (E), and sorafenib-treated (F) fli1:GFP embryos are shown. (G) Venn diagram summarizing functional categories of lead compounds in ERMS tumorigenesis. |
|
CHIR99021 inhibits ERMS tumor growth and alters differentiation of tumor cells. (A and B) Representative images of ERMS-bearing fish after 6 d of drug treatment with DMSO (A) or 400 nM CHIR99021 (B). (Scale bar, 0.2 cm.) Heat map scale indicates increasing tumor volume intensity. (C–F) H&E-stained sections (C and D) and immunohistochemistry for β-catenin (E and F). (Scale bar, 20 μm.) (G) Summary of tumor volume changes. N, number of tumor-bearing fish per treatment group. Each error bar denotes SEM. (H and I) Summary of frequency of tumor cell subpopulations upon treatment with (H) DMSO (vehicle) and (I) CHIR99021 by FACs analysis. Each pie chart represents an average of three tumor-bearing fish analyzed, showing the relative percentage of myf5:GFP+ /mylz2:mCherry tumor-propagating cells (green), myf5:GFP+/mylz2:mCherry+ (yellow), and late-differentiating myf5:GFP/mylz2:mCherry+ cells (red). SD for each fraction is indicated. *Significance of <0.05. (J) Quantitative RT-PCR analysis. *P < 0.05, Student t test. Each error bar indicates SEM. n = 3–4 tumors analyzed for each treatment group. (K and L) Immunohistochemistry for Phospho-H3 for DMSO- (K) and CHIR99021-treated (L) tumors. The number of Phospho-H3-positive cells was quantified in three independent fields at 400× magnification. The values (with SEM) indicate average of three tumors analyzed for each treatment group (P < 0.05). (Scale bar, 20 μm.) (M) Summary of EdU analysis. Larval fish engrafted with tumors expressing myf5:GFP and mylz2: mCherry were treated with DMSO or CHIR99021 and pulsed with EdU for IF analysis. Each error bar indicates SEM of three tumors from each treatment group. *P < 0.005, Student t test. (N–W) Representative images from EdU IF staining. GFP (N and O), mCherry (P and Q), DAPI (R and S), and EdU (T and U). (V and W) Merge image of all four channels. Yellow arrowheads, representative myf5:GFP+/mylz2:mCherry cells that have EDU incorporation. (Scale bar, 20 μm.) |
|
BIO alters the differentiation status of zebrafish ERM tumor cells. (A–C) Transplanted ERMS tumor-bearing larval zebrafish were treated with DMSO and BIO (300 nM) for 5 d. MF20 staining was performed on cryosections of treated tumors to assess the extent of terminal differentiation. (A) DMSO-treated tumor. (B) BIO-treated tumor. Three independent tumors from each treatment group were analyzed. Average percentage of MF20-positive cells ±SD are noted. (Scale bar, 20 μM.) (C) Real-time RT-PCR assessing expression of myogenic markers in DMSO- and BIO-treated fish (n = 3). Error bar indicates SD across experimental triplicate. *P < 0.05 in comparison with the DMSO-treatment group. Myosin heavy chain-6 (mhc). (D and E) Immunohistochemistry for Phospho-H3. The number of Phospho- H3-positive cells was quantified in three independent fields at 400× magnification. The values (with SEM) indicate average of three tumors analyzed for each treatment group. (F–O) EdU analysis. Larval fish engrafted with tumors expressing myf5:GFP and mylz2:mCherry were treated with DMSO or BIO and pulsed with EdU for immunofluorescence (IF) analysis. GFP (F and G), mCherry (H and I), DAPI (J and K), and EdU (L andM). (N and O) Merge image of all four channels. Yellow arrowheads denote representative myf5:GFP+/mylz2:mCherry cells that have EdU incorporation. (Scale bar, 50 μM.) (P) Analysis of apoptosis by Annexin V staining after multiparameter FACS. (Q) Quantitation of caspase-3 immunohistochemical staining. Average of number of positive cells in three high-power (400× magnification) fields is shown for three independent tumors treated with either DMSO or BIO. Each error bar indicates SD. NS, not significant. |