- Title
-
Gene-expression analysis of hair cell regeneration in the zebrafish lateral line
- Authors
- Jiang, L., Romero-Carvajal, A., Haug, J.S., Seidel, C.W., Piotrowski, T.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
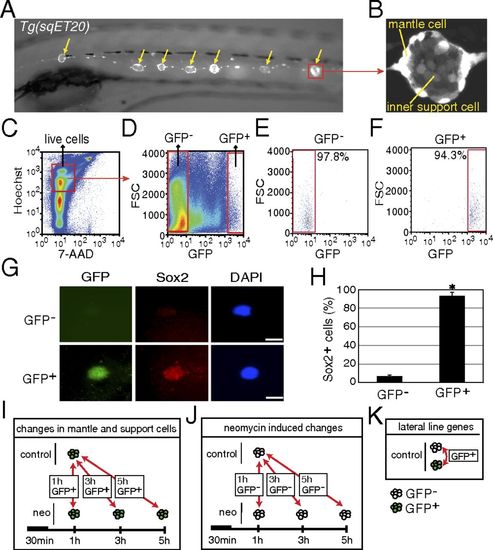
Purification of mantle and inner support cells from Tg(sqET20). (A) In 5-dpf Tg(sqET20) larvae GFP is expressed in neuromasts (arrows) and in the interneuromast cells that connect adjacent neuromasts. (B) Magnification of a neuromast shows that mantle cells are strongly labeled by GFP, but inner support cells are weakly labeled. (C and D) Dissociated cells of Tg(sqET20) larvae at 5 dpf were FACS sorted using a two-gate sorting strategy. Live cells were gated by 7-AAD and Hoechst+ (C) and subsequently were gated by forward scatter (FSC) and GFP intensity to isolate GFP and GFP+ cells (D). (E and F) A postsorting experiment shows that GFP cells are 97.8% pure (E) and GFP+ cells are 94.3% pure (F). (G) Sorted GFP and GFP+ cells were stained with Sox2 antibody and DAPI. (Upper) A GFP cell is Sox2. (Lower) A GFP+ cell expresses Sox2. (Scale bars: 5 μm.) (H) Quantification of Sox2+ cells in each population as mean ± SD; *P < 0.01 by t test. (I) Regeneration-induced mantle and inner support cell genes were identified at each time point by comparing GFP+ neomycin-treated cells with GFP+ untreated samples. (J) Neomycin-induced genes were identified by comparing GFP neomycin-treated cells at each time point with GFP untreated samples. Neomycin-induced genes subsequently were subtracted from the gene lists obtained with GFP+ cells to select for regeneration-specific changes in gene expression in inner support and mantle cells. (K) Comparisons between GFP+ and GFP cells of untreated larvae generated a list of genes enriched in lateral line cells. EXPRESSION / LABELING:
|
|
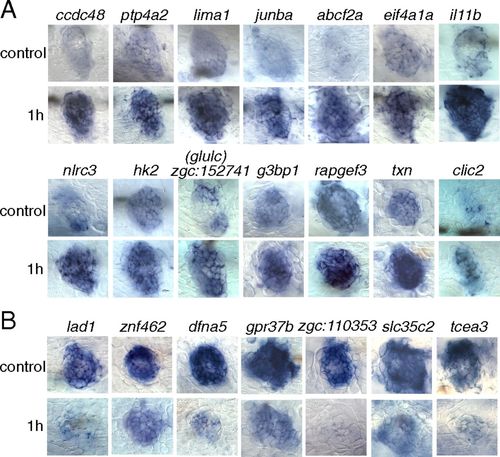
Validation by in situ hybridization of a selection of 14 genes up-regulated (A) and seven genes down-regulated (B) at 1 h after neomycin treatment. (Upper Rows) Untreated larvae (control) at 5 dpf. (Lower Rows) Treated larvae. |
|
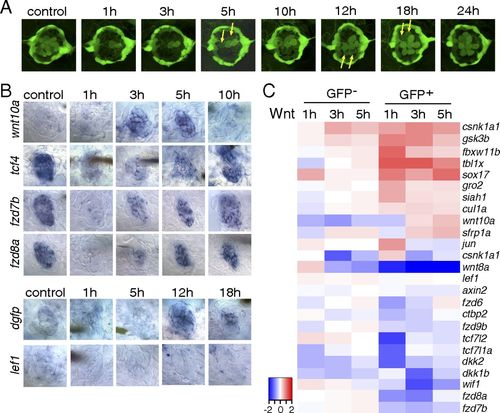
The Wnt/β-catenin pathway is inactive during the early stages of hair cell regeneration. (A) Still images of a Tg(sqET20;sqET4) larval neuromast in the process of neomycin-induced regeneration. All hair cells, except for two immature hair cells, were killed by neomycin by 1 h after neomycin treatment. Two newly formed hair cells (arrows) start to express GFP at 5 h, and other pairs of hair cells (arrows) appear at 12 and 18 h. (B) In situ hybridization of Wnt/β-catenin pathway genes in 5-dpf untreated (control) larvae and in larvae at different time points after neomycin treatment. Expression of wnt10a is increased at 3 and 5 h. Expression of tcf7l2 (tcf4), fzd7b, and fzd8a is largely decreased at 1 h after neomycin. The Wnt/β-catenin reporter line Tg(Tcf/Lef-miniP:dGFP) demonstrates activation starting at 12 h as shown by dgfp expression, but lef1 is not induced in regenerating neuromasts. (C) Heat map of selected Wnt/β-catenin pathway genes based on RNA-Seq results. Log2 ratios of GFP and GFP+ cells at 1, 3, and 5 h after neomycin treatment are color coded (red: up-regulation, blue: down-regulation). Genes that are modulated in GFP cells are neomycin-induced genes. EXPRESSION / LABELING:
|
|
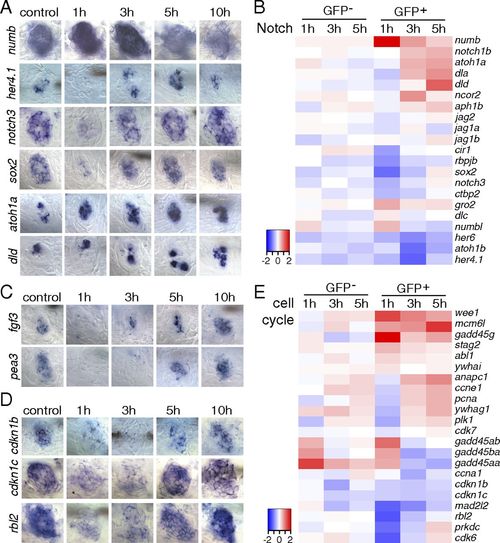
Inhibition of Notch and Fgf pathways is accompanied with cell cycle reentry immediately after hair cell ablation. (A, C, and D) In situ hybridization of Notch pathway, Fgf pathway, and cell cycle genes in 5-dpf untreated and neomycin-treated larvae. (A) The Notch pathway inhibitor numb is increased at 1 h after neomycin. The Notch target genes her4.1, notch3, and sox2 are decreased at 1 h and up-regulated at 3 h after neomycin treatment. atoh1a is up-regulated starting at 1 h after neomycin, and its target dld is increased at 5 and 10 h. (C) fgf3 and its target pea3 are down-regulated at 1 and 3 h after neomycin treatment. (D) Cell cycle inhibitory genes cdkn1b, cdkn1c, and rbl2 are decreased at 1 and 3 h after neomycin compared with control. (B and E) Heat maps of selected Notch pathway genes and cell cycle genes. |
|
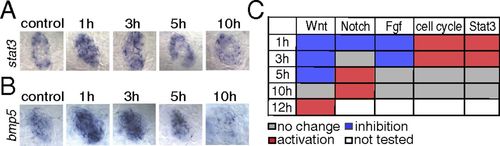
The Jak1/Stat3 pathway is activated during the early stages of hair cell regeneration. (A) In situ hybridization shows that stat3 is increased at 1 and 3 h after neomycin treatment. (B) bmp5 is up-regulated at 1 h after neomycin treatment in situ. (C) Table depicting the status of signaling pathways at different time points after hair cell death. All comparisons (no change, inhibition, or activation) were evaluated relative to untreated control samples. The Wnt, Notch, and Fgf pathways are inhibited immediately after neomycin treatment, whereas the cell cycle and Jak1/Stat3 pathways are activated during the early stages of regeneration. The Wnt and Notch pathways are reactivated at 12 h and 5 h after neomycin treatment, respectively. EXPRESSION / LABELING:
|
|
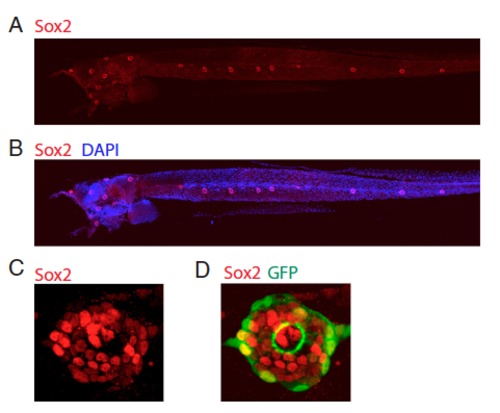
Tg(sqET20) larvae (5 dpf) stained with Sox2 antibody and DAPI. (A and B) Sox2 specifically labels neuromasts. (C and D) Magnification of a neuromast. Sox2 antibody labels inner support cells as well as some GFP+ mantle cells. |
|
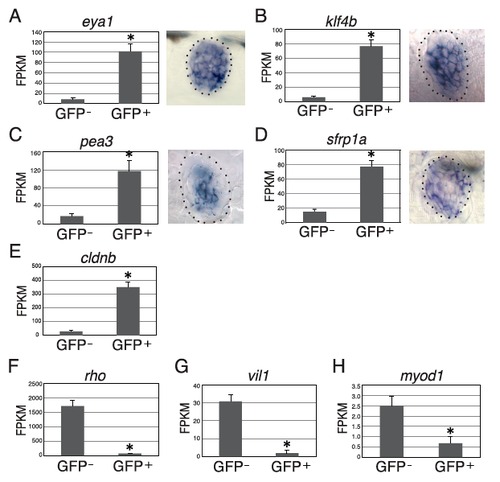
GFP and GFP+ cells from control Tg(sqET20) larvae were analyzed for gene expression by using FPKM levels from RNA-Seq. (A–D) (Right) In situ hybridization indicates that eya1 is expressed in all neuromast cells, klf4b and pea3 are expressed in inner support cells, and sfrp1a is expressed in mantle cells and some inner support cells. (A–D, Left and E) Expression of eya1 (A), klf4b (B), pea3 (C), sfrp1a (D), and cldnb (E) is increased significantly in GFP+ cells compared with GFP cells, indicating that not only mantle but also inner support cells are enriched in GFP+ cells. (F–H) The retina marker rho (F), gut marker vil1 (G), and skeletal muscle marker myod1 (H) are expressed at very low levels in GFP+ cells. All results are presented as mean ± SD; *P < 0.01 by t test. |