- Title
-
UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma
- Authors
- Mudbhary, R., Hoshida, Y., Chernyavskaya, Y., Jacob, V., Villanueva, A., Fiel, M.I., Chen, X., Kojima, K., Thung, S., Bronson, R.T., Lachenmayer, A., Revill, K., Alsinet, C., Sachidanandam, R., Desai, A., SenBanerjee, S., Ukomadu, C., Llovet, J.M., and Sadler, K.C.
- Source
- Full text @ Cancer Cell
|
High UHRF1 Expression Causes Global DNA Hypomethylation (A) 5MeC levels and total DNA stained with methylene blue were measured in 5 dpf control and UHRF1-GFP High livers (n = 4) and liverless carcasses (n = 3). The ratio of 5MeC to total DNA was averaged and normalized to controls. Student’s t test was used to determine p values. (B) Confocal stacks of livers (top) and fins (bottom) from 5 dpf nls-mCherry and UHRF1-GFP High larvae stained with anti-5MeC. Because a hepatocyte-specific promoter was used for transgenesis, there was no transgene expression in the fin. (C) Dnmt1 is uniform in the hepatocyte nucleus of four dpf nls-mCherry larvae but is found in GFP-containing punctae in UHRF1-GFP High hepatocytes. Arrows point to cells that do not express GFP and have Dnmt1 distribution pattern similar to controls. (D) Tg(hsp70I:UHRF1-EGFP) and nontransgenic controls were heat shocked at 37C for 1 hr at 24 and 27 hpf, treated with 10 μM MG132 or DMSO at 28 hpf, and collected at 34 hpf for immunoblotting. (E) Dnmt1 levels normalized to tubulin were averaged from six experiments. Student’s t test was used to determine p values; n.s., not significant; error bars represent SD. |
|
UHRF1-Induced Hypomethylation Reduces Liver Size (A) Individual larvae were imaged daily from 3–10 dpf. (B) Five dpf UHRF1-GFP High larvae display a range of liver sizes scored as ‘‘normal’’ or ‘‘small’’. (C) Three clutches were scored according to criteria in (B); n, number of larvae. Fisher’s exact test was used to determine p value. (D) The area of the left liver lobe was measured in 5 dpf fish from two clutches. Boxes represent 75th and 25th percentile, horizontal line is the median, and whiskers mark lowest and highest values. Student’s t test was used to determine p value. (E) UHRF1-GFP High larvae were sorted by liver size on 5 dpf and tracked daily for survival to 20 dpf. Data are pooled from three clutches. (F) UHRF1-GFP High and control larvae were treated with 50 mM 5-Aza from 2.5–5 dpf and scored for liver size in six clutches. Fisher’s exact test was used to determine p values. (G) UHRF1-GFP High embryos were injected with mRNA encoding dnmt1 or Mpi before 1 hpf. The percent of fish with a normal liver size was scored at 5 dpf in six clutches. EXPRESSION / LABELING:
PHENOTYPE:
|
|
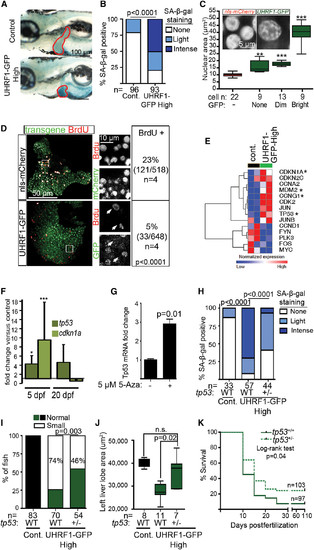
UHRF1 Overexpression in Hepatocytes Induces Tp53-Mediated Senescence (A) Intense senescence-associated β-galactosidase (SA-b-gal) staining was detected in the liver (outlined) of 5 dpf UHRF1-GFP High larvae compared to light or no staining in controls. (B) Five dpf fish from five clutches were scored for hepatic SA-b-gal staining. ***p < 0.0001 by Fisher’s exact test. (C) Nuclear size was measured in hepatocytes of a single control or UHRF1-GFP High 5 dpf liver, and cells were stratified according to GFP expression. Inset shows confocal stack of the DNA organized into foci. **p < 0.001 and ***p < 0.0001, compared to nuclear size in nls-mCherry larvae. (D) BrdU-positive cells and the total number of transgene-expressing hepatocytes in nls-mCherry and UHRF1-GFP High larvae (bottom) 5 dpf larvae. A Fisher’s exact test was used to calculate p value. In nls-mCherry larvae, most BrdU-positive cells also express the transgene, whereas the BrdUpositive cells in UHRF1-GFP High livers did not express GFP (white arrows in magnified regions, which are marked by the white box). (E) Heatmap of log2 values from RNA-seq shows cell-cycle regulators are down and Tp53 target genes (marked by *) are up in UHRF1-GFP High 5 dpf livers. (F) tp53 and cdkn1a mRNA expression were induced on 5 dpf and downregulated on 20 dpf in UHRF1-GFP High livers. *p = 0.05; ***p = 0.001 calculated by one sample Student’s t test. Error bars represent SD. (G) 5-Aza induces Tp53 expression in primary mouse hepatocytes. Student’s t test was used to determine p value with SD indicated by the error bars across three replicates. (H–K) tp53+/- in UHRF1-GFP High larvae significantly reduced SA-b-gal staining in the liver (two clutches) (H), and increased the percent of larvae with normal liver size (I), the area of the left liver lobe (J), and survival at 5 dpf (K). p values were calculated with a Fisher’s test with Freeman- Halton extension (H), Fisher’s exact test (I), and Student’s t test. Boxes represent 75th and 25th percentile, horizontal line is the median, and whiskers mark lowest and highest values (J). |

UHRF1 Is an Oncogene (A) Atypical cells, dysplastic foci (outlined), and HCC are apparent in hematoxylin and eosinstained UHRF1-GFP High livers. BD, bile duct; MF, mitotic figure. (B) Incidence of normal and atypical hepatocytes, dysplastic foci, and cancer in the liver of UHRF1- GFP High fish on WT or tp53+/- background. (C) NIH 3T3 cell growth in soft agar is enhanced when UHRF1 overexpression is combined with RAS (n = 3). The p value was calculated by Student’s t test, and error bars represent the SD. (D) Hepatic SA-b-gal-staining patterns in UHRF1- GFP High larvae change as fish age. Images of 8 dpf larvae illustrate the SA-b-gal-staining patterns that were scored in the time course shown in the graph. A significant increase in the number of UHRF1-GFP High fish with intense or punctate SA-b-gal compared to controls at all time points (p < 0.01 by Fisher’s exact test) except at 4 dpf; n.s., not significant. (E) BrdU incorporation in the liver on 11 dpf is five times higher in UHRF1-GFP High fish than in controls. Total number of cells counted is indicated with n = number of clutches assessed. Fisher’s exact test was used to calculate p value. EXPRESSION / LABELING:
PHENOTYPE:
|
|
UHRF1 mRNA and Protein Are Overexpressed in HCC (A) UHRF1 detected by qRT-PCR in 18 preneoplastic lesions and 40 HCCs from hepatitis C virus (HCV)-infected patients compared to expression in nine normal livers. Horizontal line indicates median. (B) Immunohistochemistry for UHRF1 protein (brown) was evaluated in 52 of the same HCCs examined in (A) plus five normal liver samples. Fisher’s exact test was used to calculate p value. Seventy-one of the HCV-associated HCCs analyzed by qPCR were grouped into high (n = 35) and low (n = 36) UHRF1-expressing tumors based on the median log2-fold change of 3.64. (C) HepG2 cells transfected with control siRNA (GL2) or two different siRNAs targeting UHRF1 described in Tien et al. (2011) were blotted for UHRF1 and cleaved and total PARP (arrow indicates full length; * indicates cleaved protein). (D–H) Vascular invasion (33 high and 34 low tumors; four missing values) (D), serum AFP (29 UHRF1-high and 29 low tumors; 13 missing values) (E), early (<2 years) (F) and late (>2 years; 32 UHRF1-high and 35 low tumors; four missing values) (G) tumor recurrence, and overall survival after surgery (32 high and 35 low tumors; four missing values) (H) were stratified according to UHRF1 expression. Continuous and categorical variables were assessed by Wilcoxon rank-sum test and Fisher’s exact test, respectively. Clinical outcome difference was evaluated by log rank test. In box and whisker plots, boxes represent the 75th and 25th percentiles, the whiskers represent the most extreme data points within interquartile range 3 1.5, and the horizontal bar represents the median. |
|
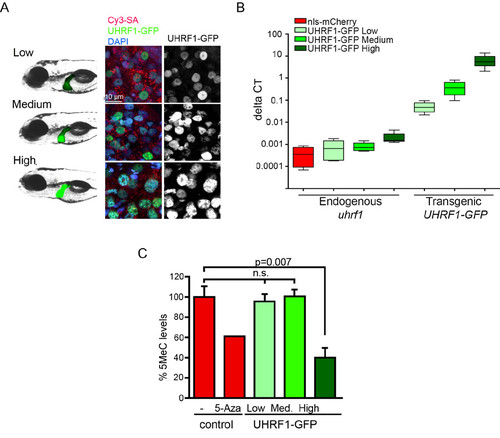
DNA hypomethylation is specific to UHRF1-GFP High larvae. (Related to Figure 1) (A) Three independent F1 zebrafish lines over-expressing UHRF1 at different levels were generated. Live images (left) and confocal sections through the liver (right) stained with Cy3-SA (red) and DAPI (blue) from 5 dpf transgenic fish confirm the nuclear localization of the transgene and reveal variable levels of expression. With increasing levels of UHRF1-GFP, nuclei become larger and DNA becomes punctate. (B) Endogenous (zebrafish) uhrf1 and transgenic (human) UHRF1 mRNA levels in the liver of each transgenic and control line (nlsmCherry) was determined by qPCR on 5 dpf. Four separate clutches of pooled livers were analyzed per line. Boxes represent 75th and 25th percentile, bars indicate lowest and highest values. (C) Global DNA methylation determined fromslot blots of genomic DNA from the liver of 5 dpf controls (Tg(fabp10:nls-mCherry or non-transgenics)) that were either untreated or treated with 50 μM 5-Aza (n=1) and from Tg(fabp10:UHRF1-GFP) larvae expressing Low, Medium and High levels of UHRF1 in the liver (n=3). Samples blotted in parallel were stained with either methylene blue for total DNA or probed with anti-5MeC. The 5MeC signal was normalized to methylene blue staining. Average, normalized values were divided by the average value in control (untreated livers). p value was determined by Student’s T-test; not significant (n.s.) Error bars are SD. |

Only high UHRF1 expression leads to a reduced liver size and increased larval death. (Related to Figure 2) (A) Cy-3 streptavidin stained the entire liver and then the area of the left liver lobe of at least 5 fish from each UHRF1-GFP transgenic line and from control fish on 3-5 dpf was measured. Liver size was only significantly reduced in UHRF1-GFP High fish on 5 dpf as calculated by a Student’s T-test. (B) Standard length of nlsmCherry and UHRF1-GFP High fish on 5 and 10 dpf. (C) Three clutches of each of the UHRF1 over expressing lines (High, Medium and Low) and control fish ((Tg(fabp10:dsRed) or Tg(fabp10:nls-mCherry)) were monitored for survival from 5-500 dpf. A significant decrease in survival by log-rank test was found only for UHRF1-GFP High and UHRF1-GFP Medium lines. (D) UHRF1-GFP High embryos were injected with mRNA encoding zebrafish Dnmt1 or mannose phosphate isomerase as a control (Chu et al., 2013) prior to the 8 cell stage. On 5 dpf, fish were fixed and stained with Cy3-SA to label the liver. The area of theleft liver lobe was calculated for at least 10 fish per condition. Uninjected Tg(fabp10:nls-mCherry) and UHRF1-GFP High larvae were also measured as controls. Percent of liver size was normalized to the median liver size in control, uninjected larvae and indicated on the graph. Boxes represent 75th and 25th percentile, bars indicate lowest and highest values. Error bars are SD. |
|
UHRF1 overexpression in hepatocytes induces senescence independent of apoptosis. (Related to Figure 3) (A) TUNEL assay was used to assess apoptosis in uhrf1 mutant fish and UHRF1-GFP High fish. Apoptotic cells were seen in 5 dpf uhrf1 mutants, but not in the livers of UHRF1-GFP High fish. Images were taken at 10x. (B) Senescence associated β-galactosidase (SA-β-gal) staining in the liver was scored for each of the three UHRF1-GFP transgenic lines and control larvae on 5 dpf. The number of fish scored and the number of clutches (in parenthesis) are indicated. |
|
Histological features of dysplasia and HCC caused by UHRF1 over expression. (Related to Figure 4) (A) H&E sections of UHRF1-GFP High livers in 20 dpf fish illustrate features of dysplasia and HCC that are not present in nls-mCherry controls. Features represented in the images in A include hepatocyte crowding, ductular reaction (DR), presence of mitotic figures (MF), atypical cells (atyp) and enlarged senescent cells (indicated by white arrows). The two areas marked by * in the far left panel of UHRF1-GFP High fish indicate two separate lesions in the sameliver. The boxed region is enlarged in the panel on the right. (B) Histological criteria including cytological and architectural changes used to define features of atypical cells, dysplastic foci and HCC in zebrafish. (C) PCNA positive and negative hepatocytes were counted in >200 hepatocytes from 2-8 fish per cohort of Tg(fabp10:nls-mCherry) and UHRF1-GFP overexpressing lines at 20 dpf and 40 dpf. Error bars are SD. (D) 20 dpf Cherry controls and UHRF1-GFP High sections were stained with anti-5MeC and the percent of 5MeC stained hepatocytes is shown. Arrows point to positively staining cells that are not hepatocytes. (E) Two UHRF1-GFP High fish were randomly selected on 20 dpf to image GFP in the liver as a measure of transgene expression. (F) qRT-PCR analysis of UHRF1 mRNA expression at 5 and 20 dpf normalized to rpp0 demonstrates that transgene expression is maintained, albeit decreased, in older UHRF1-GFP High fish. Boxes represent 75th and 25th percentile, bars indicate lowest and highest values. |
Reprinted from Cancer Cell, 25(2), Mudbhary, R., Hoshida, Y., Chernyavskaya, Y., Jacob, V., Villanueva, A., Fiel, M.I., Chen, X., Kojima, K., Thung, S., Bronson, R.T., Lachenmayer, A., Revill, K., Alsinet, C., Sachidanandam, R., Desai, A., SenBanerjee, S., Ukomadu, C., Llovet, J.M., and Sadler, K.C., UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma, 196-209, Copyright (2014) with permission from Elsevier. Full text @ Cancer Cell