- Title
-
A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos
- Authors
- Bhatia, S., Monahan, J., Ravi, V., Gautier, P., Murdoch, E., Brenner, S., van Heyningen, V., Venkatesh, B., and Kleinjan, D.A.
- Source
- Full text @ Dev. Biol.
|
Characterisation of a novel conserved element E+120 in mouse and zebrafish reporter transgenics. (A) The E+120 element (orange ellipse) is located 120 kb 3′ of the PAX6 P1 promoter in the human locus, in the large final intron of ELP4. (B) VISTA plot using the human sequence as a base, showing the deep conservation of E+120 in mouse, chicken, coelacanth, one of the zebrafish pax6 loci and the elephant shark. The constructs used for the reporter transgenic experiments are shown. (C) Four independent transgenic mouse lines were analysed for the E120Z construct, showing variable patterns of expression at E11.5. (D) At E17.5 strong expression is seen in the cerebella (black arrow) and olfactory bulbs (white arrowhead) in 3 out of 4 lines. (E) Expression pattern of the elephant shark E+120 element in transgenic zebrafish, shown by mCherry (CHR) fluorescence. Signal is seen in the olfactory placodes (op) and olfactory bulbs (ob) from 24 hpf. At 48 hpf (shown at two different confocal plains) and 96 hpf expression is also found at the midbrain-hindbrain boundary (mhb) and in the hindbrain (hb) region. |
|
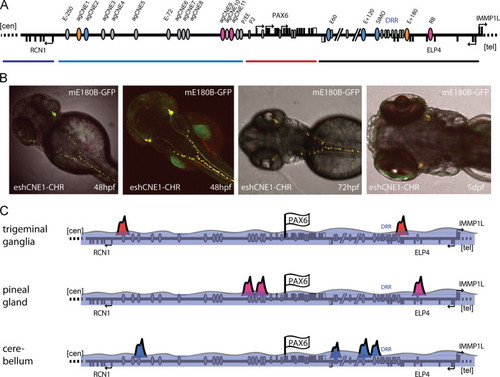
Characterisation of the E+180 conserved element. (A) Overview of the PAX6 downstream region indicating the position of the E+180 element at the telomeric end of the DRR. (B) The E+180 element contains three peaks of sequence conservation between human, mouse, chicken, coelacanth and elephant shark, but no significant conservation is seen in the zebrafish pax6 loci. A schematic overview of putative cis-elements in the telomeric half of the DRR shows the location of the E+180 elements and the fragments used in the transgenic reporter experiments. (C)–(H) Reporter transgenic mice with the E180 fragment from elephant shark (E180A/B/C). Strong expression is present at E13.5 in the trigeminal ganglion (TG) and in the dorsal root ganglia (DRG) along the neural tube (NT). At E13.5 expression also appears in the ganglion cell layer (GCL) of the eye (E) and in the optic nerves (ON). At E17.5 staining is present in the olfactory bulbs (OB) and olfactory epithelium (OE), in nerve tracts along the neural tube and hindbrain, and remains strong in the trigeminal ganglion and nerves. Expression is also present in the retinal ganglion cells, and staining extends along the optic nerve, chiasm (OC) and optic tracts (OT) to the lateral geniculate nuclei (LGN). (I)–(Q) Transgenic embryos with the mouse E180B only fragment. Reporter expression first becomes apparent from E11.5 in the trigeminal ganglion and increases in strength towards E13.5. Expression in the ganglion cell layer of the retina and in the optic nerve starts around E13.5 and continues to manifest at E17.5 in the retinal ganglion cells, optic nerve, chiasm and optic tracts and lateral geniculate nuclei. Staining is also seen in the olfactory bulbs and epithelium. (R)–(U) The mouse and elephant shark E180 fragments were also tested in reporter transgenic zebrafish. In situ hybridisation with a GFP antisense probe on mE180B-GFP transgenic fish shows the expression in the trigeminal nerve and tracts along the neural tube. Dual colour fluorescence in mouse E180B-GFP and elephant shark E180-mCherry double transgenic fish show the near-complete overlap in expression. |
|
Characterisation of tissue-specific expression patterns of CNEs from the RCN1 to PAX6 intergenic region. (A) Schematic overview of the region with the positions of the ancient gnathostome CNEs indicated by ellipses. The agCNEs (orange) and the chick-mammalian specific CNEs (purple) characterised in this figure are highlighted. (B) Representative reporter transgenic embryos are shown for agCNE1-5 and for the less deeply conserved E-250 and E-72 elements. Only background signal is seen with the E-250 and E-72 fragments. agCNE1 drives expression in the trigeminal ganglia and dorsal spinal cord neurons. agCNE2 transgenic embryos show signal in forebrain, olfactory bulbs and placodes and hindbrain. Expression from agCNE3 is seen in the olfactory placodes and olfactory bulbs with weak expression in hindbrain in some of the lines. In contrast agCNE4 shows strong hindbrain specific expression. agCNE5 shows consistent expression in the forebrain, with hindbrain expression in some lines. |
|
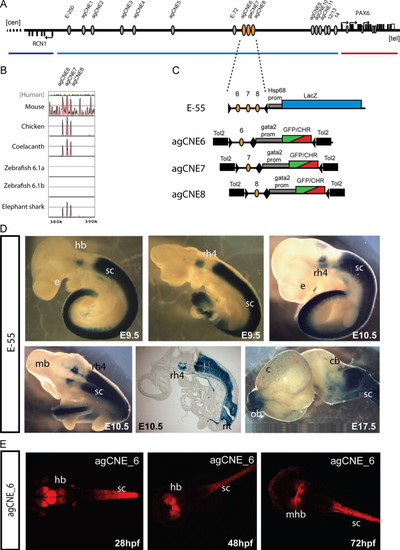
Functional dissection of the E-55 region. (A) Schematic overview of the RCN1 to PAX6 intergenic region with the positions of agCNEs indicated by grey ellipses and agCNE6-8 in orange. (B) VISTA plot of the multispecies sequence alignment of the fragment showing the three peaks of conservation in elephant shark but not in either of the zebrafish pax6 loci. (C) Schematic depiction of the reporter constructs used to generate transgenic mice and fish. (D) Transgenic embryos for the E-55 fragment, containing all three agCNEs, show X-gal staining in rhombomere 4 (rh4) of the hindbrain (hb) and from rhombomere 7 towards caudal along the length of the spinal cord, covering its full area on cross-section (sc). Expression in the neural tube is maintained at E17.5 and also seen in the olfactory bulbs (ob) in some embryos. (E) Transgenic reporter embryos carrying agCNE6 only show restricted expression in part of the hindbrain (hb), particularly at the midbrain-hindbrain boundary (mhb) and along the length of the spinal cord (sc). EXPRESSION / LABELING:
|
|
Characterisation of anciently conserved elements in the Pax6 promoter and proximal region. (A) VISTA plot of multispecies sequence conservation in the PAX6 genomic region containing the promoters and immediate upstream CNEs. PAX6 exons are shown as blue peaks and non-coding peaks of conservation are in pink. The positions of the validated Pax6 promoters P0, P1 and Pα are indicated, as are the known enhancers P/EE and Panc2 corresponding to agCNEs 12-14. agCNEs 9-11 are well conserved in all vertebrate Pax6 loci analysed with the exception of agCNE11 which is absent in the zebrafish pax6b locus. Tracks underneath the plot show the presence of several CpG islands in the region, including one at the agCNE9-10 location, and a simplified overview of transcripts produced from the region. In addition to the various Pax6 transcripts two transcripts are found on the opposite strand, the multi-exon Pax6OS and the 2.8 kb single exon ncRNA AK032637. (B) rtPCR analysis of AK032637 in comparison with Pax6 and Gapdh in a selection of cell lines and embryo tissues. N2A, neuroblastoma, bTC3, pancreatic β-cell line, MV+, lens epithelium, RAG, renal adenocarcinoma, vTh, ventral thalamus, eye, E17.5dpc whole eye, mb, midbrain, ctx, cortex. (C) Reporter transgenic zebrafish with construct agCNE10-mCherry show ubiquitous low level signal but lack specific expression. (D) Transgenic fish for agCNE9-GFP show expression in the pineal gland with additional signal in the fore- and hindbrain in some lines. Fluorescent signal in transgenic fish with the agCNE11-GFP construct is restricted to the pineal gland only. |
|
Multiple CNEs with overlapping expression patterns form enhancer archipelagos at the PAX6 locus. (A) Overview of the locus with newly characterised agCNEs depicted as ellipses. CNEs with overlapping regulatory activities found in this study are indicated with matching colours. (B) Overlap in the expression patterns of the agCNE1 and E180B enhancers is shown by dual-fluorescence in double transgenic zebrafish at 24 hpf, 48 hpf and 72 dpf. (C) Schematic depiction of the inter-digitated co-existence of multiple regulatory archipelagos at the PAX6 genomic locus. Selected archipelagos are activated in a tissue-specific manner by the binding of TFs and cofactors at defined CNEs. |

Table S2: Overview of ancient gnathostome conserved elements (agCNEs) characterised in this study. The table shows the agCNE, the species of origin of the element, the model organism and the reporter gene used for the transgenic assay, followed by the number of independent expressing lines analysed, with the total number of transgenics shown in brackets for the mouse experiments. The final column shows the sites of reporter expression with the number of lines showing the pattern described in brackets. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
Reprinted from Developmental Biology, 387(2), Bhatia, S., Monahan, J., Ravi, V., Gautier, P., Murdoch, E., Brenner, S., van Heyningen, V., Venkatesh, B., and Kleinjan, D.A., A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos, 214-228, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.