- Title
-
Targeted inactivation and identification of targets of the Gli2a transcription factor in the zebrafish
- Authors
- Wang, X., Zhao, Z., Muller, J., Iyu, A., Khng, A.J., Guccione, E., Ruan, Y., and Ingham, P.W.
- Source
- Full text @ Biol. Open
|
Expression of Hedgehog target genes in wild-type and gli mutant embryos. ptch2, prdm1a, nkx2.2a and olig2 were examined in WT (A–D), gli1ts269/ts269 (E–H), gli2ai276/i276 (I–L), gli1ts269/ts269;gli2ai276/i276 (M–P), gli2ai276/ty119 (Q–T), and gli2aty119/ty119 (U–X) by WISH. All specimens were fixed at 24hpf, except those hybridized with prdm1a probe (18 somites). EXPRESSION / LABELING:
|
|
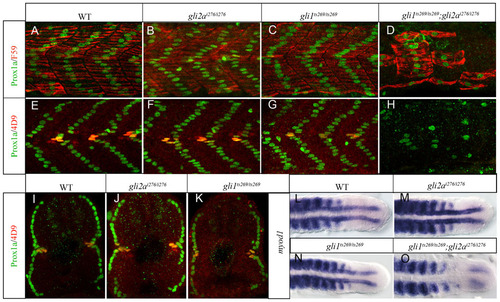
Gli1 and Gli2a act redundantly to pattern the zebrafish myotome. (A,E) Wild-type (WT); (B,F) gli2ai276; (C,G) gli1ts269 and (D,H) gli1ts269; gli2ai276 24hpf embryos stained with anti Prox1a (green) and either mAb F59 (red: panels A–D) or 4D9 (red: panels E–H) to reveal slow-twitch muscle and MP fibres. (I–K) Transverse sections of embryos shown in panels E–G. (L–O) Dorsal views of the caudal regions of 18ss embryos hybridized with a probe for myod1 transcript. (L) WT; (M) gli2ai276; (N) gli1ts269 (O) gli1ts269;gli2ai276. EXPRESSION / LABELING:
PHENOTYPE:
|
|
The Gli1 and Gli2a activators differ in their ability to counter Gli2a-mediated repression. (A,B,E,F) gli1ts269/+; gli2aty119/+ (C,G) gli2ai276/ty119 (D,H), and gli1ts269/+; gli2ai276/ty119 (D) embryos at 24hpf stained with anti Prox1a (green) and either mAb F59 (red: panels A–D) or 4D9 (red: panels E–H) to reveal slow-twitch muscle and MP fibres. All panels show the caudal somites; the white dotted line indicates the position of the end of the yolk sac extension. The somites shown in panels C and G are located at 3–5 somites away from the end of the yolk sac extension. (I–L) Expression of myod1 in (I) gli2ai276/ty119, (J) gli1ts269/+; gli2aty119/+, (K) gli1ts269/+; gli2ai276/ty119, and (L) gli2aty119/ty119 embryos at 18 ss. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Gli bound region on ptch1 and ptch2 genomic locus. (A) The consensus Gli binding site (GBS). (B) Profile of Gli2a ChIP-seq sequence enrichment at the ptch1 genomic locus. (C) Predicted GBS located within each peak (I–IV) and their conservation between different species. (D) Profile of Gli2a ChIP-seq sequence enrichment at the ptch2 genomic locus. (E) Predicted GBS located within each peak (I–III) and their conservation between different species. (F) Transient expression of GFP driven by a wild-type 8kb ptch2 promoter fragment that includes the Peak I GBS. (G) Transient expression of the same reporter in which the GBS is mutated. |
|
Gli bound region on gli1 and olig2 genomic locus. (A) Profile of Gli2a-ChIP sequence enrichment at the gli1 genomic locus. (B) Profile of Gli2a-ChIP sequence enrichment at the olig2 genomic locus. (C) Predicted GBS located within each gli1 peak (I–III) and their conservation between different fish species. (D) Predicted GBS located within the olig2 peak (CNE) and its conservation between different fish species. (E–G) Expression of olig2 transcript in wild-type (E1,E2), gli2aty119 (F1,F2) and dnPKA injected (G1,G2) embryos at the 15 somite stage. Left panels are dorsal view of mesoderm and right panels are section view. (H) Transient expression of GFP driven by a 1.9kb fragment including the CNE (I) transient GFP expression driven by the same 1.9kb fragment in which the GBS was mutated. EXPRESSION / LABELING:
|
|
Validation of new Gli2a targets by WISH. (A–H) Expression of seven newly identified Gli2a direct targets plus prdm8b was analysed by WISH in wild-type, gli2aty119 and dnPKA injected embryos at the 15 somite stage. Each pair of panels shows a dorsal view of mesoderm of a flat mounted embryo and a transverse section of a similar stage embryo. EXPRESSION / LABELING:
|
|
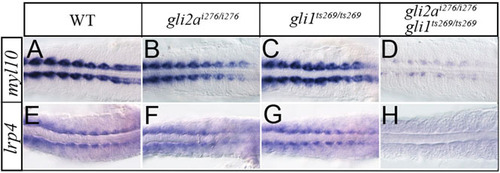
New Gli2a targets are redundantly controlled by Gli1 and Gli2a. Expression of myl10 and lrp4 expression in wild-type (A,B), gli2ai276 (C,D), gli1ts269 (E,F), and gli1ts269; gli2ai276 (G,H) 18 somite stage embryos. EXPRESSION / LABELING:
|
|
Sequence information and morphological phenotype of new gli2a mutant alleles. (A) Sequence information of gli2ai275/+and gli2ai276/+at ZFN target site. (B–G) Morphological phenotype of wild type, gli2ai276/i276, gli1ts269/ts269, gli1ts269/ts269;gli2ai276/i276, gli2aty119/ty119, and gli2ai276/ty119 at 30 hpf. (H) Alignment of Gli2a and Gli2ai276 amino acid sequence. Sequences highlighted in yellow show the common amino acids between them, sequence in red indicates the antigen region for generating the Gli2a antibody used in this study. Sequences underlined indicate the Gli2a repressor domain (blue), zinc finger DNA binding domain (pink), nuclear localization sequences (black) and activator domain (green). All domains were annotated by alignment with the human GLI2 protein (Fernandez-Zapico, 2008). |
|
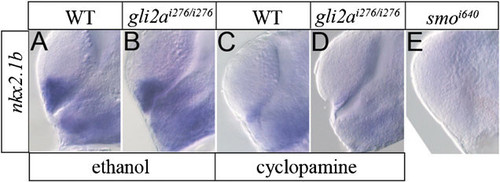
nkx2.1b expression is not changed in gli2ai276/i276. nkx2.1b expression at diencephalon and telencephalon was examined by WISH on wild type (A), gli2ai276/i276 (B), wild type treated with cyclopamine (C), gli2ai276/i276 treated with cyclopamine (D), and smoi640 (E). |