- Title
-
miR-219 Regulates Neural Precursor Differentiation by Direct Inhibition of Apical Par Polarity Proteins
- Authors
- Hudish, L.I., Blasky, A.J., and Appel, B.
- Source
- Full text @ Dev. Cell
|
Loss of Apical Polarity Correlates with Loss of Neural Precursors and Lumen Morphogenesis All images show representative transverse sections at the level of the trunk spinal cord with dorsal up. (A and B) At 1 and 2 dpf, ZO-1 is concentrated at apical membranes of cells lining a primitive lumen, which extends across the dorsoventral axis of the spinal cord (brackets). (C and D) At 3 and 5 dpf, the primitive lumen is replaced with a ventrally positioned central canal marked by apically localized ZO-1. (E–H) F-actin is similarly localized to apical membranes lining the primitive lumen and central canal. Additionally, most cells that express Sox2 are associated with F-actin localization. (I–L) A BrdU pulse labels numerous cells lining the primitive lumen at 1 and 2 dpf, but at 3 and 5 dpf few cells incorporate BrdU. Scale bar equals 10 μm. |
|
miR-219 Knockdown Causes Retention of Embryonic Spinal Cord Precursors into Postembryonic Stage All images show representative transverse sections through trunk spinal cord with dorsal up. (A–C) In situ hybridization to detect the mature form of miR-219 reveals low expression at 1 dpf but prominent expression at 2 dpf. Staining appears most intense in cells (asterisks) lateral to ventricular zone cells (vz, bracket) lining the primitive lumen. At 3 dpf, miR-219 expression appears uniform in medial spinal cord. Boxed areas are shown in higher magnification in (A2)–(C2). (D and E) miR-219 (3 dpf) MO-injected larva has more cells that incorporate BrdU than a control larva. (F) Graph showing the number of BrdU+ cells in the ventral, dorsal, and entire spinal cord at 3 dpf. Data represent the mean ± SEM (n = 10 larvae, five to ten sections each). ****p < 0.0001, unpaired t test. (G and H) miR-219 (3 dpf) MO-injected larva has more PH3+ cells than a control larva. (I) Graph showing the number of spinal cord cells labeled with anti-PH3 antibody in control and miR-219 MO-injected larvae. Data represent the mean ± SEM (n = 10 sections obtained from 15 larvae per group, with two replicates). p = 0.0328, unpaired t test. (J–L) miR-219 (3 dpf) MO-injected larvae have more Sox2+ cells than control larvae. Graph showing the number of Sox2+ cells in ventral, dorsal, and entire spinal cord. Data in graph represent mean ± SEM (n = 10 sections obtained from 15 larvae per group, with three replicates). p = 0.0369, ****p < 0.0001, unpaired t test. (J, K, M, and N) miR-219 (3 dpf) MO-injected larvae maintain a primitive lumen marked by apically localized F-actin and ZO-1 (brackets). Boxed areas are shown at higher magnification in (J2), (K2), (M2), and (N2). Scale bar equals 10 μm for low-magnification images and 5 µm for high-magnification images. See also Figure S1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
miR-219 Is Required for Differentiation of Glia and Late-Born Neurons All images show representative transverse sections through trunk spinal cord with dorsal up. (A and B) Whereas numerous GFAP+ radial glia occupy the spinal cord of a 3 dpf control larva, a miR-219 MO-injected larva has few radial glia except for the most dorsal and ventral regions of spinal cord (brackets). Asterisks mark oligodendrocyte lineage cells. (C and D) Images showing a deficit of BLBP+ radial glia in a miR-219 MO-injected larva compared to control. (E and F) miR-219 MO-injected larvae appear to have a normal number and distribution of neurons, marked by Elavl3 expression, but fewer oligodendrocyte lineage cells (asterisks) than control larvae. (G) Graph showing number of Isl+ motor neurons in control and miR-219 MO-injected larvae. Data are presented as mean ± SEM (n = 10 sections obtained from 15 larvae per group, with two replicates). p > 0.05, unpaired t test. (H and I) Confocal images of embryos pulsed with EdU at 1 dpf and fixed at 2 dpf. Numerous Edu+ cells are also Elavl3+ (arrowheads) in control larvae (H), whereas most EdU label persists within cells lining the spinal cord lumen, and fewer neurons are labeled by EdU in miR-219 MO-injected larva (I). (J) Graph showing number of Elav3+ EdU+ neurons in control and miR-219 MO-injected larvae. Data represent mean ± SEM (n = 10 sections obtained from five larvae per group). p < 0.0001, unpaired t test. Scale bar equals 10 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
miR-219 Has Single, Conserved Target Sites within prkci and pard3 32 UTRs (A and B) Schematic representations of prkci and pard3 transcripts with predicted miR-219 target sites conserved among various species. (C–E) In controls at 1 and 2 dpf, Prkci protein is concentrated at apical membranes lining the primitive lumen, but by 3 dpf Prkci is limited to the central canal (brackets). (F and G) Prkci labeling persists along a primitive lumen extending across the spinal cord dorsoventral axis in 3 dpf and 5 dpf miR-219 MO-injected larvae. (H) Sequences (220 bp) from the pard3 32 UTR containing wild-type and mutated miR-219 target sites were cloned into dual luciferase vectors. (I) Quantification of light units revealed a miR-219-mediated reduction of reporter gene expression that was abrogated by 1 and 2 bp mutations within the target site. Data represent ± SEM (three independent experiments). Brackets indicate pairwise comparisons. p < 0.0001, unpaired t test. Scale bar equals 10 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
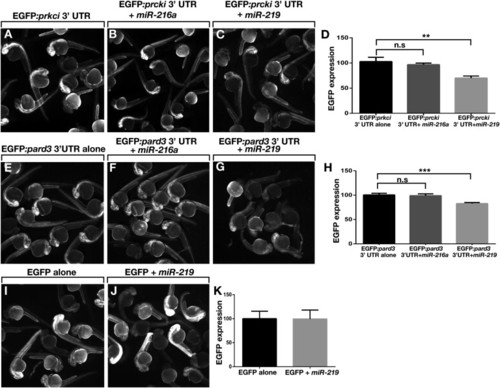
mir-219 Regulates Reporter Gene Expression In Vivo via pard3 and prkci 32 UTR Sequences (A–C) Fluorescence images of living embryos injected with EGFP:prkci 32 UTR mRNA alone, miR-216a control, or miR-219. (D) Graph showing EGFP fluorescence intensity values. Units represent pixel intensity and are reported as percent of control values (n = 20 embryos, with three replicates). Brackets indicate pairwise comparisons. p = 0.0014, unpaired t test. (E–G) Images of living embryos injected with EGFP:pard3 32 UTR mRNA alone, miR-216a control, or miR-219. (H) EGFP fluorescence intensity values shown as in (D) (n = 20 embryos, with three replicates). p = 0.0001 unpaired t test. (I and J) Images of embryos injected with EGFP mRNA alone or with miR-219. (K) EGFP fluorescence intensity values shown as in D (n = 20 embryos, with two replicates). p = 0.8728, unpaired t test. Error bars represent ± SEM. |
|
pard3 and prkci Are Functionally Relevant miR-219 Targets (A–C) Lateral images of living 3 dpf (Tg:olig2:EGFP) larvae, focused on the trunk spinal cord. Control larva (A) shows the normal number and distribution of dorsally migrating OPCs (arrow). Whereas miR-219 MO-injected larvae have few OPCs (B), larvae coinjected with miR-219 and pard3 MOs have an intermediate number of OPCs (C). (D) Graph showing quantification of OPC phenotypic classes. Larvae classified as normal had the number of dorsally migrated OPCs typical of wild-type. Larvae were classified as severe if fewer than five OPCs had migrated and mild in all other circumstances. p value was calculated by comparing the number of larvae with normal numbers of OPCs in the miR-219-MO alone and miR-219 MO ± pard3 MO experiments. Data represent ± SEM (n = 25 larvae per group, with three replicates). p = 0.0324, unpaired t test. (E and F) Larvae injected with pard3 TP MO have fewer OPCs than those injected with a control TP MO. (G) Graph showing quantification of the pard3 TP MO phenotypes. Data represent ± SEM (n = 35 and 55 larvae in two independent experiments). p < 0.0001, unpaired t test. (H and I) Larvae injected with prkci TP MO have fewer OPCs than those injected with a control TP MO. (J) Graph showing quantification of the prkci TP MO phenotype. (n = 45–60 larvae per experiment, with three independent experiments). p = 0.0020, unpaired t test. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Blocking miR-219 Access to pard3 or prkci 32 UTRs Phenocopies miR-219 Loss of Function All panels show representative images of spinal cord transverse sections with dorsal up. (A–C) Larvae (3 dpf) labeled to detect Prcki localization. In the control, Prkci is limited to a small central canal (A). By contrast, Prkci extends dorsally in pard3 TP MO and prkci TP MO-injected larvae, marking a primitive lument (B and C). (D–F) Similarly to Prcki, ZO-1 is restricted to a central canal in the control larva (D) but extends more dorsally in pard3 TP MO and prkci TP MO-injected larvae (E and F). (G–I) pard3 TP MO and prkci TP MO-injected larvae have fewer medial spinal cord radial glia than control. (J–L) pard3 TP MO and prkci TP MO-injected larvae have more Sox2+ cells in dorsal spinal cord than control. (M–O) BrdU incorporation at 3 dpf. TP MO-injected larvae have more labeled cells than control. (P) Graph showing number of BrdU+ cells in dorsal, ventral, and entire spinal cord. Data represent ± SEM (n = 10 larvae for each experiment, five to ten section per larva). p < 0.05, p < 0.005, p < 0.0005, unpaired t test. Scale bar equals 10 µm. See also Figure S2. EXPRESSION / LABELING:
PHENOTYPE:
|
|
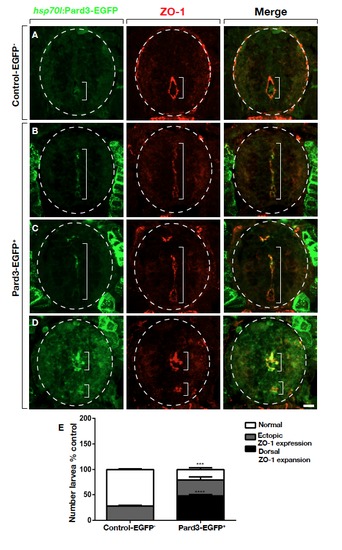
Pard3-EGFP Overexpression Phenocopies miR-219 Loss of Function Representative spinal cord transverse images of 3 dpf heat-shocked control and heat-shocked Tg(hsp70l:pard3-EGFP) larvae processed for ZO-1 immunohistochemistry with dorsal to the top. (A) Control non-transgenic larva. ZO-1 is restricted to the central canal. (B and C) Examples of heat-shocked transgenic larvae showing strong Pard3-EGFP expression outside the spinal cord (outlined) and co-localized with ZO-1 to an apparent primitive lumen extending into dorsal spinal cord. (D) Example of a heat-shocked larva with an apparent double lumen (ectopic class) with co-localized Pard3-EGFP and ZO-1. (E) Graph showing number of larvae having dorsal expansion or ectopic co-localization of Pard3-EGFP (n=12 each for control and experimental) ***P=0.0002, ****P<0.0001. Scale bar equals 10 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Cell, 27(4), Hudish, L.I., Blasky, A.J., and Appel, B., miR-219 Regulates Neural Precursor Differentiation by Direct Inhibition of Apical Par Polarity Proteins, 387-398, Copyright (2013) with permission from Elsevier. Full text @ Dev. Cell