- Title
-
Heartbeat-Driven Pericardiac Fluid Forces Contribute to Epicardium Morphogenesis
- Authors
- Peralta, M., Steed, E., Harlepp, S., González-Rosa, J.M., Monduc, F., Ariza-Cosano, A., Cortés, A., Rayón, T., Gómez-Skarmeta, J.L., Zapata, A., Vermot, J., and Mercader, N.
- Source
- Full text @ Curr. Biol.
|
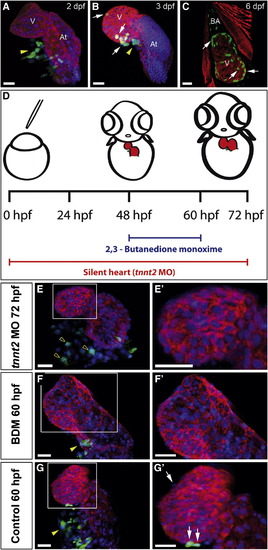
Epicardium Formation Is Inhibited in the Absence of a Heartbeat (A–C and E–G′) Immunohistochemical analysis of proepicardial (PE) and epicardial development. Panels show 3D projections of whole-mount hearts from Epi:GFP zebrafish larvae at different developmental stages stained with anti-myosin heavy chain (MHC) to detect myocardium (red) and DAPI to counterstain nuclei (blue). DAPI is not shown in (C) to improve visualization of the myocardium. GFP expression is shown in green. Yellow arrowheads mark PE cell clusters; white arrows mark epicardial cells. (E′)–(G′) are zoomed views of boxed areas in (E)–(G).(D) Scheme of experiments for inhibiting heart function by tnnt2MO-injection into single-cell stage embryos or BDM treatment between 48 and 60 hpf. Myocardium is shown in red, and GFP-positive cells are shown in green. (E) Heart of a tnnt2 morphant at 72 hpf. Empty arrowheads mark scattered GFP-positive mesothelial cells at the venous pole, which do not form a PE cluster (Movie S1). (F) Heart of a BDM-treated larvae. (G) Control at 60 hpf. At, atrium; BDM, 2,3-butanedione monoxime; dpf, days postfertilization; hpf, hours postfertilization; tnnt2 MO, troponin t2 morpholino; V, ventricle. Scale bars represent 30 μ m. See also Figure S1 and Movie S1. |
|
The Heartbeat Is Necessary for PE Cluster Formation and Correct Transfer to the Myocardium Panels show experiments performed with Epi:GFP larvae. (A–D′) Confocal imaging of hearts at different stages. Dorsal is on the top. (A and A′) In wild-type larvae at 55 hpf (control), mesothelial cells of the pericardial wall (white arrows) form a PE cluster (yellow arrow) (Movie S2). (B and B′) BDM treatment inhibits PE formation and PE cells flatten (white arrow) instead of rounding up. (C–D′) Epicardial cells proliferate normally in the absence of a heartbeat. Red arrowheads mark cells about to proliferate; blue arrowheads mark newly formed cells. (E and E′) Quantification of proliferation (pH3 staining) and apoptosis (TUNEL staining) of pericardial mesothelial cells in the presence or absence of a heartbeat; 3 and 5 hr BDM treatments from 60 hpf onward were compared with controls fixed at the same stage. The y axis shows number of pericardial cells. p < 0.001. (F and F′) Pericardial cavity volume at 48 and 72 hpf in systole (S) and diastole (D) in untreated larvae (n = 2 and 3) and after BDM treatment initiated 1 or 12 hr before, as indicated (n = 4 and 6). (G–H′) Larvae with an altered heartbeat show a different pattern of myocardial ventricle colonization. (G and G′) A few epicardial cells (yellow arrowhead) are observed in tnnt2 morphants with <100% penetrance, which have a weak heartbeat and strong edema. The white and blue dotted lines outline the pericardial cavity and myocardium, respectively. (H and H′) Order of myocardial colonization by PE cells. In control larvae, epicardial cells first colonize the distal ventricle surface (D), while in weak-heartbeat tnnt2 morphants epicardial cells mostly colonized the proximal ventricle (P). Numbers indicate the percentages of animals with the reported colonization pattern and the total numbers of animals analyzed. (I–I′′) Enlarged PE cluster in caffeine-treated animals. (I) PE cell quantification of caffeine-treated (n = 16) versus untreated larvae (n = 16) revealed significant differences (p = 0.023; two-tailed Student’s t test). (I′ and I′′) Merged bright-field and fluorescent images of PE clusters (yellow arrows) from a control and a caffeine-treated larva. The arrowhead indicates epicardial cells. (J and J′) Impaired PE and epicardium formation in myh6 morphants revealed by cell quantification at 70 hpf (n = 21 control and 45 myh6 morphants; p < 0.0001, two-tailed Student’s t test). In all graphs, bars indicate means and standard deviations. D, distal; myh6MO, myosin heavy polypeptide 6 morpholino; ns, not significant; P, proximal; pH3, phospho-Histone 3. Scale bars represent 10 μm (A–D′, I, and I′) and 20 μm (G and G′). See also Movie S2. |
|
Proepicardial Cells Are Released from Three Distinct Sources (A) Confocal imaging of a 48 hpf Epi:GFP heart showing GFP-positive PE clusters at the atrioventricular canal (avcPE) and venous pole (vpPE) (Movie S3). (A′) Location of the PE cells shown in (A)–(D): advected PE cells (PEC), avcPE cells, vpPE cells, and epicardial progenitors arising from the arterial pole pericardial mesothelium (apEP). The white arrow indicates direct transfer from the pericardial mesothelium to the myocardium; red arrows indicate release into the pericardial cavity (gray shading). (B) apEP cells transfer directly to the myocardium. At the onset of acquisition (0 ms), the apEP cell (blue dotted circle) is attached to the pericardial wall. (B′) Later (2,284 ms), the apEP cell has transfered to the myocardial surface (Movie S4). (B′′) Tracking of an apEP cell. Note the limited movement in response to contact with the ventricle before transfer (blue trace). Red lines mark the regions in which displacement was measured for kymographs. Motion after transfer is comparable to that of an epicardial cell attached at an earlier time point (red trace). (C and C2) A vpPE cluster (black dotted circle) moving in response to heart contraction, which leads to its release (PE cluster reveals a limited motion before release and increased oscillatory motion afterward, coupled to atrial wall contraction. (D and D′) A pair of advected PE cells (PEC white dotted circles) in the cavity circumnavigate the ventricle in the direction denoted by the yellow arrow (Movie S6). (D′′ and D′′′) Kymograph analysis and highlighted representation showing coupling of this motion to atrial contraction. In all kymograph analyses, displacement values enable the synchronization of PE cell motion after attachment to the myocardium or release from the cluster to be seen. In all panels, gray arrows mark epicardial cells on the myocardial surface prior to acquisition and asterisks indicate PE cell positions before release. Scale bars represent 20 μm. See also Movies S3, S4, S5, S6, and S7. |
|
Characterization of Flow Forces within the Pericardial Cavity (A and A′) Measurement of the distance between the myocardium and the pericardial wall during heart contraction. (A) Ventral view of a 48 hpf heart immersed in Bodipy TR. The continuous and discontinuous lines outline the myocardium at diastole and systole, respectively; ventricle is in green, and atrium is in yellow. (A′) Average maximum distance over four heartbeats between the pericardial wall and ventricle (green) and pericardial wall and atrium (yellow). (B) Localization of PE cells within three regions (R1, R2, and R3) of the pericardial cavity. Numbers are the percentages of cells within each region from a total of 30 cells or cell clusters. (C and D) Tracking of a PE cell (dot) at the periphery of the pericardial cavity (C) and tracking of a PE cell flowing around the ventricle (D) (Movie S8). Speed (μm/s) of advected PE cells is represented by a color code; minimum and maximum speeds are indicated. (E) Speed of advected cells within the pericardial cavity (means ± SD), varied significantly in different areas, indicated by letters a–d (one-way ANOVA followed by Tukey’s mixed-effect test. p < 0.05 for all pairwise comparisons). (F) Positions probed with optical tweezers: 1, cell positioned close to the vpPE site; 2, cell close to the avcPE site; 3, cell at a distance from the ventricle; 4, cell close to the ventricle; and 5, cell positioned at the AVC. (G and H) Vector and force maps obtained by optical tweezing. (I–K′) In vivo confocal imaging and tracking of fluorescent beads injected into the pericardial cavity; beads recapitulate the pattern of advected PE cells (Movie S9). (K and K′) Advected PE cell (K) and bead (K′) following the same flow. (L) Model summarizing the circulatory pattern of PE cells moving with the pericardial flow in conjunction with the observed order of myocardial adhesion. PEC, proepicardial cells; pN, picoNewton. Scale bars represent 50 μm. See also Movies S8 and S9. |
|
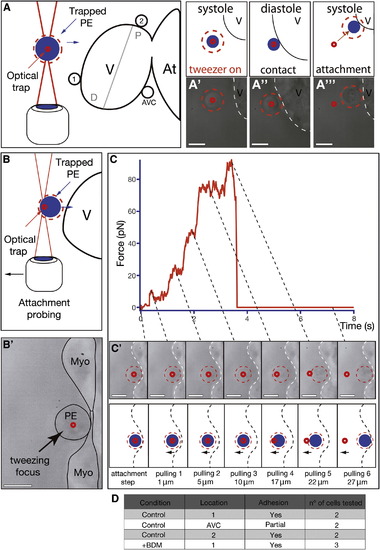
Characterization of PE Cell Adhesion to the Myocardium (A) Optical-tweezers setup used to study the propensity of PE cells to adhere to different myocardial regions including it, a region distal to it (region 1), and a region proximal to the AVC (region 2). The blue circle illustrates the trapped cell and the small red circle the approximate location of the optical trap. The red dotted circle around the cell facilitates its visualization during the experiment. (A′–A′′′) Trapped PE cells adhere firmly to the ventricle if placed close to it (Movie S10). (B–C′) Optical-tweezers pulling test to determine the role of heartbeat during PE cell attachment to the myocardium. After BDM treatment, a PE cell was trapped and forced to attach to the myocardium (time 0 s, B2 and C). After 30 s, the trapped cell was progressively pulled away from the myocardium. (C and C′) Optical-tweezers forces applied to a cell over time during the pulling test. With increased distance, the magnitude of the force within the tweezers increases (10 pN, 20 pN, 40 pN, 70 pN, 90 pN), eventually resulting in the cell moving out of the tweezers because of its attachment to the myocardium (pulling 5). Further pulling resulted in the cell springing back to its original position on the surface of the heart (pulling 6) (Movie S11). The dotted lines link the force changes observed in the trap (C) with the images of the trapped cell at the same time (C′). (D) Table summarizing the number, location, and types of adhesion events tested with the optical tweezers. Scale bars represent 10 μm. See also Movies S10 and S11. |
|
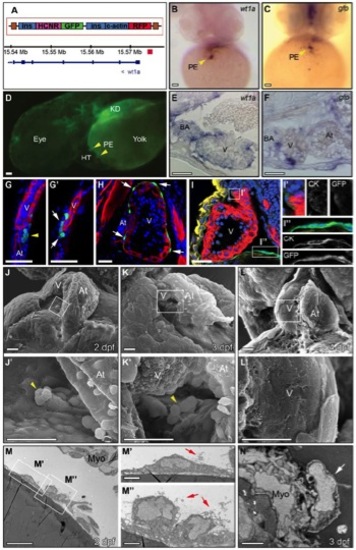
Epicardium Development Visualized by a Zebrafish Reporter Line and by Electron Microscopy, Related to Figure 1 |