- Title
-
Laminin beta1a controls distinct steps during the establishment of digestive organ laterality
- Authors
- Hochgreb-Hägele, T., Yin, C., Koo, D.E., Bronner, M.E., and Stainier, D.Y.
- Source
- Full text @ Development
|
s804 mutants display defects in left-right asymmetry of endodermal organs. (A,A′) The liver (Li) and pancreas (Pa) in wild-type and s804 mutants are revealed by GFP expression in the enhancer trap line Tg(XlEef1a:GFP). Ventral views, anterior towards the top. (B,B2) myl7 and foxa3 expression in wild-type (B) and s804 mutants (B2), as revealed by whole-mount in situ hybridization. myl7 is expressed in cardiomyocytes and foxa3 is expressed in the gut primordium. Dorsal views. Dashed lines mark the midline. (C) Repartition of embryos according to cardiac asymmetry, as determined by myl7 expression. The numbers of embryos analyzed are indicated at the bottom. (D,D′) Confocal imaging projection of the liver (Li) and pancreas (Pa) at 72 hpf in wild-type (D) and s804 mutants (D′), as revealed by Tg(XlEef1a:GFP) expression (green). Ventral views. Islet 1 (red); pan-cadherin (blue). Dotted lines outline islet1-positive endocrine cells in the principal islet and throughout the hepatopancreatic ductal system. Scale bars: 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
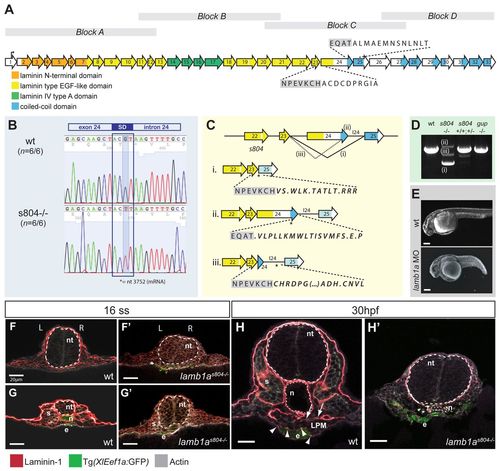
Mutation in a splice donor (SD) site of the lamb1a gene results in a truncated protein in s804 mutants and impairs expression of the multimeric protein laminin 1 in the basement membrane of lamb1as804 mutants. (A) The lamb1a ORF in zebrafish (exons are numbered). Functional domains of the corresponding protein are represented by colored boxes. Gray blocks represent the cloning strategy. (B) G-to-T replacement in the SD site for exon 24 of lamb1a in s804 mutants (6/6). (C) Alternative splicing in lamb1as804 mutants. Mutation of the SD site for exon 24 results in excision of exon 24 (i), maintenance of intron 24 (I24, ii) or splicing of exon 23 into a cryptic site within exon 24 (iii). These alternative splice events result in a frame-shift and premature stop signals (asterisks) that disrupt the C-terminal coiled-coil domains (blue boxes). (D) PCR amplification of block C from wild-type cDNA produces a single band of 1.5 kb. In s804 mutants, alternative products are detected. (E) Injection of a lamb1a splice-blocking MO recapitulates the s804 mutant phenotype. Scale bars: 20 μm. (F-H′) Transverse sections of wild-type (F,G,H) and lamb1as804 mutant (F′,G′,H′) embryos at 16 ss (F-G′) and 30 hpf (H,H2). Anterior (F,F′) and posterior (G,G′) level sections. Lam1 (red), Tg(XIEef1a:GFP) (green). L, left; R, right. Scale bars: 20 μm. Neural tube (nt, broken line), notochord (n), somite (s) and gut endoderm (e). Dotted line identifies marked atrophy of the notochord. EXPRESSION / LABELING:
|
|
Lamb1a plays multiple roles in the establishment of left-right asymmetry. (A,B) Establishment of L-R gene expression machinery is examined by expression of southpaw/spaw in wild-type (A) and lamb1as804 mutant (B) embryos at 21 ss. (C,D) Integrity of the notochord is assessed by expression of no tail/ntl at 21 ss. Reduction of ntl expression in lamb1as804 mutants is restricted to the level of the visceral organ-forming region (white bracket). (A-D) Dorsal views. (E-H) Single-plane confocal imaging of wild-type embryos stained for Lam1 (green), acetylated tubulin (red) and phalloidin (blue). Dorsal views, anterior towards the top. Expression of Lam1 during KV morphogenesis in the epiblast (E) and at the margin (F) of 80% epiboly stage embryos. Discrete foci of Lam1 deposition are associated with the lumen of the vesicle (G, arrowheads) and where cilia formation takes place (H, arrows). Scale bars: 10 μm. (I,J) Confocal imaging projection of KV cilia at 8 ss, as revealed by acetylated tubulin staining (red) in wild-type (I) and lamb1as804 mutant (J) embryos. Counterstaining by phalloidin (blue). Scale bars: 10 μm. Dorsal views, anterior towards the top. (K,L) Quantitative analysis (mean±s.e.m.) of KV cilia length in wild type and lamb1as804 mutants. Asterisks indicate statistical significance, *P<0.0001 and **P=0.031. (M-O) Projection of bead trajectories depicts the fluid flow in the KV of wild-type (M) and lamb1as804 mutant (N,O) embryos at 10-12 ss. Lines depicting trajectories of beads within the KV and speed (μm/s) shown are for a representative embryo. (P) Quantitative analysis (mean±s.e.m.) of KV fluid flow in wild-type and lamb1as804 mutant embryos. Results depict average KV flow speed in six wild-type and six lamb1as804 mutant embryos. Asterisks indicates statistical significance, ***P=0.022. EXPRESSION / LABELING:
PHENOTYPE:
|
|
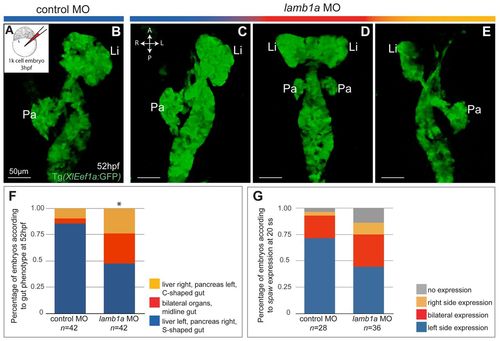
Knockdown of lamb1a in the KV results in randomization of liver and pancreas asymmetry. (A-E) Confocal imaging of the gut in Tg(XlEef1a:GFP) embryos injected at 3 hpf (A) with control (B) or lamb1a (C-E) MO. Effect of KV knockdown of lamb1a on the L-R asymmetry of the liver (Li) and pancreas (Pa) was analyzed at 52 hpf. Resulting phenotypes of the liver and pancreas after KV-specific lamb1a knockdown include: wild type (C, blue); bilateral liver and pancreas and midline gut (D, red); inversion of L-R asymmetry, with the liver and pancreas localized to the right and left sides of the embryo, respectively (E, yellow). Ventral views. (F) Percentages (mean±s.e.m.) of embryos displaying distinct liver and pancreas L-R asymmetry phenotypes. Forty-two embryos were analyzed for each treatment. Asterisks indicate statistical significance, *P=0.001. (G) Percentages (mean±s.e.m.) of embryos showing distinct spaw expression patterns at 20 ss. Wild-type embryos were injected with control (n=28) or lamb1a (n=36) MO at 3 hpf. |
|
Loss of Lamb1a function affects LPM morphogenesis essential for asymmetric LPM migration and gut looping. (A,B) Expression of Tg(hand2:EGFP) in the LPM of wild-type (n=5) and lamb1as804 mutant (n=5) embryos at 25 hpf. GFP (green), phalloidin (red). (C-G) Four classes of LPM migration phenotypes were observed in wild-type (n=10) and lamb1as804 mutant (n=24) embryos at 30 hpf. In G, the arrow indicates the aberrant protrusion of a Tg(hand2:EGFP)-expressing cell into the gut. The arrowhead indicates a Tg(hand2:EGFP)-expressing cell located inside the gut. (A′-G′) The LPM and gut shown in A-G. Gut (yellow); left LPM (red); right LPM (blue); Tg(hand2:EGFP)-expressing cells (green). (H) Proportions of wild-type (n=10) and lamb1as804 mutant (n=24) embryos exhibiting the four classes of LPM migration phenotypes; *P<0.001. (I) Ratios (mean±s.e.m.) of Tg(hand2:EGFP)-expressing cells to Tg(hand2:EGFP)-non-expressing cells in the LPM at 25 and 30 hpf. Five wild-type and five mutants were examined at 25 hpf. Ten wild-type and 24 mutants were examined at 30 hpf. Asterisks indicate statistical significance: **P<0.05; ***P<0.01. (J,J2) Expression of the LPM marker wnt2bb in wild-type (J) and lamb1as804 mutant (J2) embryos at 30 hpf. Arrows indicate the expression in the LPM at the gut-looping region. Dorsal views, anterior towards the top. (K,K2) Expression of the tight junction protein ZO-1 (red) and Tg(hand2:EGFP) (green) in the LPM of wild-type (K) and lamb1as804 mutant (K2) embryos at 32 hpf. Arrows in K2 indicate LPM cells that protrude into the gut but still maintain localized ZO-1 expression. (A-G,K,K2) Transverse sections, dorsal towards the top. Scale bars: 10 μm. Broken lines outline the LPM. Shadows in K and K′ outline the gut primordium. L, left; R, right. |
|
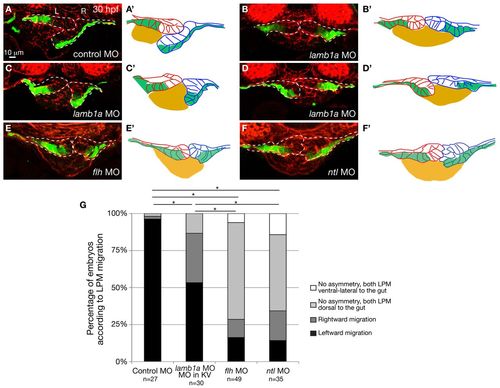
lamb1a knock-down in the KV as well as global knock-down of flh and ntl randomized the direction of LPM migration without altering epithelial polarity or composition of the LPM. (A-F) Expression of Tg(hand2:EGFP) in the LPM of embryos injected with control (A) or lamb1a (B-D) MO into the KV at 3 hpf, or flh MO (E) or ntl MO (F) at the one-cell stage. Transverse sections, dorsal towards the top. Scale bar: 10 μm. Dashed lines outline the LPM. L, left; R, right. (A′-F′) The LPM and gut shown in A-D. Gut (yellow), left LPM (red), right LPM (blue), Tg(hand2:EGFP)-expressing cells (green). (G) Proportions of control, lamb1a, flh and ntl MO-injected embryos exhibiting different directions of LPM migration. Effect of lamb1a knockdown in the KV on LPM migration is statistically significant, when compared with control, flh and ntl morphants (*P<0.001). The distribution of LPM migration phenotype is similar in flh and ntl morphants (P=0.794), but significantly different in embryos injected with lamb1a MO in the KV or with the control MO. |