- Title
-
Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis
- Authors
- Choudhuri, A., Maitra, U., and Evans, T.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
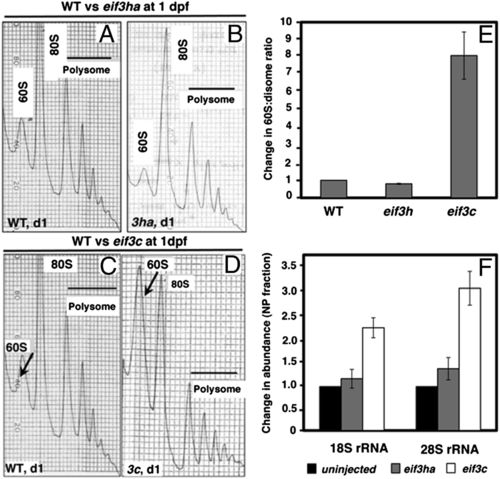
Comparison of the polysome profiles obtained from cell-free extracts of eif3ha or eif3c morphant embryos with the corresponding WT embryos. (A-D) Representative profiles derived from cell-free extracts of WT or morphant embryos, as indicated. The arrows in C and D indicate changes in the amounts of 60S subunits observed in the stage-matched WT and eif3c morphants at 1 dpf. (E) Quantitation showing the change of the 60S:disome ratios for individual eif3 morphants with respect to the corresponding WT embryos. (F) qPCR data measuring the relative abundance of 18S rRNA and 28S rRNA, as surrogate measures for the changes in abundance of 40S and 60S subunits, respectively. PHENOTYPE:
|
|
Validation of the change in translation state for genes identified through RNA-seq. Shown are qPCR results from three independent biological isolates of both WT and eif3ha morphants for 25 genes that had been found to be decreased in translational state (TS) and five randomly chosen genes that had shown no change in TS. The bars represent the mean and the error bars the SEM. The change in TS has been calculated using the formula [(WT/MO)poly/[(WT/MO)total], where (WT/MO)poly and (WT/MO)total represent the changes of individual mRNAs in polysomal and total RNA, respectively. A negative sign is added arbitrarily to the calculated values to indicate that the translation efficiency is reduced in the morphants relative to WT. Actual data for each independent set, compared with the RNA-seq data, are presented in Table S2. |
|
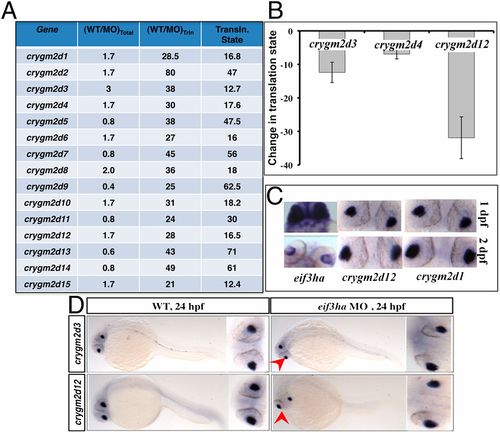
Crystallins of the crygm2d family are a cohort of genes that are translationally regulated by eif3ha. (A) Transcripts encoding crygm2d isoforms are shown according to their respective changes in the translation state relative to total RNA. (B) The change in translational state was validated in independent experiments by qPCR for crygm2d3, crygm2d4, and crygm2d12. (C) Representative in situ hybridization experiment showing that crygm2d1, crygm2d12, and eif3ha transcripts are colocalized in the developing lens at 1–2 dpf. (D) Representative in situ hybridization experiment showing that transcript patterns for crygm2d3 and crygm2d12 are unaltered in eif3ha morphants at 24 hpf compared with stage-matched WT controls. |
|
Polysome profile analysis and the strategy of subsequent RNA-seq of polysome-associated mRNAs in zebrafish embryos. (A) A cell-free extract prepared from actively translating cells is loaded onto a linear 10–50% sucrose gradient and subjected to velocity gradient centrifugation. Free 40S, 60S, and 80S ribosomes, in addition to polysomes, are size fractionated from the translationally inactive mRNAs and messenger ribonuclear protein (MRNP) particles. The A254 profile is analyzed with an attached UV-absorbance monitor. The position and integrity of the ribosomal components in the separated fractions are demonstrated by isolating total RNA from individual fractions and analyzing aliquots by gel electrophoresis. The presence of 18S, 28S, or equimolar 18S and 28S ribosomal RNAs (rRNAs) identifies the positions of 40S, 60S, and 80S ribosomes, respectively. Polysomal fractions also yield both 18S and 28S rRNAs and consist of translating mRNAs. (B) To identify genes that are translationally regulated, stage-matched WT and morphant embryos were collected, and whole-cell lysates were processed as in A. Both total RNA and polysome-associated RNA were used to generate cDNA libraries that were analyzed by deep sequencing to identify transcripts underrepresented in the morphant samples (in the polysome, but not the total RNA samples). Here we compared eif3ha and control samples at 24 hpf. (C) Flowchart indicating the protocol optimized to obtain reproducible polysome profiles from cell-free extracts of zebrafish embryos. (D) A typical polysome profile prepared from fresh (nonfrozen) WT embryos. (E) A representative polysome profile prepared from frozen WT embryos. The positions of 40S, 60S, and 80S ribosomes are indicated. |
|
Deregulated transcripts are expressed in the organ systems that are consistent with the spatial expression patterns and morphant phenotypes of eif3ha morphants. Shown is a Database for Annotation, Visualization and Integrated Discovery (DAVID) gene ontogeny analysis of gene sets deregulated from polysomes in the eif3ha morphant embryos. Shown below are the in situ hybridization patterns showing the spatial expression of eif3ha at 24 hpf indicating a correlation between the tissue association of the transcripts and the expression domains (see also ref. 2). |
|
The approach used to compare the synthesis of Crygm2d7 protein from the corresponding FLAG-tagged crygm2d7 mRNA in stage-matched control and eif3ha morphant embryos. (A) Strategy used to generate mRNA by in vitro transcription encoding C-terminally Flag-tagged Crygm2d7. In an alternate construct, the FLAG tag was placed at the N terminus of the protein. (B) The purified RNA was injected along with either a control or eif3ha-specific MO, followed by Western blotting experiments of the embryo-derived lysates ~24 h later, using anti-FLAG antibody. |
|
In situ hybridization experiments demonstrating the spatial expression patterns of ectopically injected crygm2d7-FLAG mRNA. The crygm2d7 UTR sequences are not sufficient to confer eif3ha dependency for translation. (A) Embryos were probed using an anti-crygm2d7 RNA probe. Shown are representative 24 hpf embryos that had been injected with the crygm2d7-FLAG mRNA or stage-matched control uninjected embryos, as indicated (note that transcripts in the controls are restricted to the lens). (B) The renilla luciferase (RLUC) cDNA was flanked by the crygm2d7 UTR sequences, and activity relative to firefly luciferase (FLUC) was measured in eif3ha and control morpholino-injected embryos. FLUC was used as control luciferase in this dual luciferase assay. Results shown are the mean from three independent experiments. |