- Title
-
Cftr controls lumen expansion and function of Kupffer's vesicle in zebrafish
- Authors
- Navis, A., Marjoram, L., and Bagnat, M.
- Source
- Full text @ Development
|
Generation of a cftr mutant zebrafish. (A) A TALEN was designed to target the EcoRV site in the sixth transmembrane domain. The spacer is marked by green text and the restriction site is denoted by red text. (B) Schematic of the domain structure of Cftr with a star indicating the TALEN target site within the third transmembrane domain of the protein. NBD, nucleotide binding domain; R, regulatory domain. (C) TALEN activity generated insertions and deletions that disrupt the EcoRV site. (D) Sequence alignments of the TALEN-generated alleles. Cftrpd1048 has a six-nucleotide deletion, cftrpd1049 and cftrpd1050 have nucleotide insertions and deletions causing frameshifts leading to premature stop codons. (E) An alignment of the amino acid sequences encoded by cftrpd1049 and WT cftr. |
|
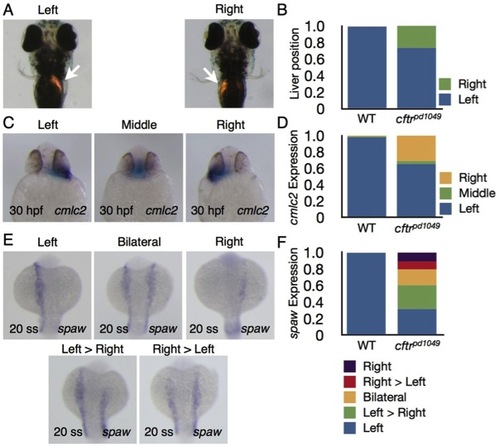
Organ laterality is disrupted in cftr1049 mutant embryos. Heterozygous cftrpd1049 fish were mated to assess organ laterality. (A) Ventral view of 4 days post-fertilization (dpf) WT and cftrpd1049 mutant larvae expressing dsRed in the liver (arrows). (B) Quantification of liver orientation in WT and cftrpd1049 mutants. Liver orientation is reversed in 27% of homozygous mutants. WT, n=103; cftrpd1049, n=30. (C) Ventral view of representative cftrpd1049 mutants showing cmlc2 expression pattern. (D) Quantification of heart looping in WT and cftrpd1049 mutants. WT, n=408; cftrpd1049, n=138. (E) Dorsal view of cftrpd1049 mutant embryos displaying left, right, left>right, right>left and left=right spaw expression patterns. (F) Quantification of spaw expression in WT and cftrpd1049 mutants. WT, n=146; cftrpd1049, n=73. EXPRESSION / LABELING:
PHENOTYPE:
|
|
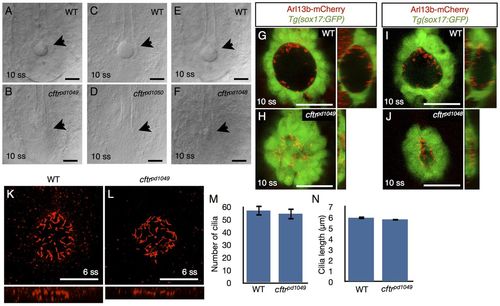
KV lumen inflation requires cftr. (A-F) KV lumen (arrow), imaged at 10 ss by DIC microscopy in WT (A,C,E) and mutant siblings (B,D,F respectively). (B,D) The KV lumen is undetectable by DIC in cftrpd1049 and cftrpd1050 mutants at 10 ss. (F) DIC imaging of the cftrpd1048 tailbud identified a small KV lumen. (G-J) Live confocal images showing ventral cross-section and orthogonal views of WT and mutant KV expressing Tg(sox17:GFP) and Arl13b-mCherry RNA at 10 ss in (G,H) WT and cftrpd1049 mutant siblings and (I,J) WT and cftrpd1048 siblings. (K,L) Confocal images of WT and mutant KV stained for acetylated tubulin. (M) Quantification of the number of KV cilia. WT KV contained 57.0±4.1 cilia (n=5) and mutant KV contained 54.5±4.6 cilia (n=8) (P=0.69). (N) Quantification of cilia length in WT and mutant KV. WT cilia length was 5.97±0.13 μm (n=30) and cftrpd1049 cilia length was 5.82±0.13 μm (n=49) (P=0.42). Error bars represent s.e.m. Scale bars: 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Apical-basal polarity is not affected in cftrpd1049 mutants. (A,B) Confocal image and associated orthogonal projections of WT and cftrpd1049 mutant fish stained for ZO-1 and Cadherin. (A′-A′′ ′,B′-B′′ ′) Magnification of the KV epithelium to show localization of the polarity markers. (C,D) Confocal image and associated orthogonal projections of WT and cftrpd1049 mutant fish stained for aPKC and expressing Tg(sox17:GFP). (E,F) Confocal image of WT and cftrpd1049 mutant KV expressing apically localized, GFP-tagged Podocalyxin. (G,H) Live confocal images of WT and cftrpd1049 mutant fish co-expressing an apical marker, RFP-Clic5b, and Tg(sox17:GFP). Scale bars: 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
cftr expression is enriched in KV. (A) At 3 ss, in situ hybridization detects cftr expression specifically in KV (arrow). (B) In situ hybridization showing cftr expression in KV (arrow) and the chordamesoderm in 10 ss embryos. (C) Schematic of the BAC recombineering procedure, showing the recombination target and the expected structure of the resulting GFP fusion protein. (D,F) DIC images showing ventral (D) and lateral (F) views of 10 ss embryos expressing TgBAC(cftr-GFP). The arrow marks the characteristic KV structure. (E,G) Whole-mount epifluorescence of the embryos shown in D and F demonstrate specific KV expression of Cftr-GFP (arrows) at 10 ss. Scale bars: 50 μm. EXPRESSION / LABELING:
|
|
Cftr is apically localized in KV epithelial cells throughout its morphogenesis. (A) Live, time-lapse confocal imaging of Cftr-RFP in TgBAC(cftr-RFP); Tg(sox17:GFP) embryos. Cftr-RFP is expressed and apically localized in KV throughout the initial stages of lumen coalescence. (B) Merge of the RFP and GFP channels. (C,D) Live confocal imaging of TgBAC(cftr-RFP); Tg(sox17:GFP) embryos at 10 ss shows that Cftr-RFP is localized apically in KV. (D) Merged view of Cftr-RFP and GFP. (E,F) Live confocal imaging of TgBAC(cftr-GFP) embryos injected with membrane-RFP RNA shows continued apical localization of Cftr-GFP until 15 ss. The dashed line marks the edge of KV. Scale bars: 50 μm. EXPRESSION / LABELING:
|
|
Fluid secretion regulates KV size. (A,B) DIC imaging of KV (arrowheads) in (A) DMSO control and (B) 10 μM forskolin-, 40 μM IBMX-treated embryos. (C) The KV area of DMSO and treated fish is quantified. DMSO n=15; forskolin + IBMX, n=13; *P<0.01. (D,E) DIC imaging of KV (arrowheads) in (D) DMSO control and (E) 1 μM ouabain-treated embryos at 10 ss. (F) Quantification of KV area in DMSO-and ouabain-treated embryos. DMSO, n=10; ouabain, n=10; *P<0.02. (G-I) Live confocal imaging of Tg(sox17:GFP)-expressing fish treated with (G) DMSO, (H) ouabain or (I) forskolin and IBMX. Error bars represent s.e.m. Scale bars: 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression of Cftr-GFP can rescue lumen expansion defects in cftr mutants. (A-C) Representative DIC images of KV (arrows) at 10 ss in embryos (A) heterozygous for cftrpd1049, (B) homozygous for cftrpd1049 and (C) in cftrpd1049 homozygous mutants injected with 150 pg cftr-GFP RNA. (D) Quantification of KV phenotype at 10 ss in control and cftr-GFP-injected embryos resulting from a cross between cftrpd1049 heterozygous parents. Control, n=158; Cftr-GFP, n=94; P<0.01. (E) Graph of the KV phenotype at 10 ss in embryos from a cftrpd1049/+; TgBAC(cftr-GFP) × cftrpd1049/+ cross, compared by whether the embryos were Cftr-GFP positive or negative. Cftr-GFP negative, n=140; Cftr-GFP positive, n=149; P<0.001. (F) Quantification of KV phenotype in embryos treated with DMSO (n=96) or 10 μM forskolin and 40 7mu;M IBMX resulting from a cross between cftrpd1049 heterozygous parents. n=95; P=0.6374. Scale bars: 50 μm. PHENOTYPE:
|
|
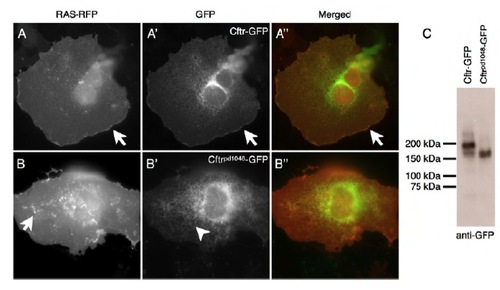
Cftrpd1048-GFP is mislocalized. (A-A′′) Zebrafish Cftr-GFP expressed in Cos-7 cells is predominantly localized to the plasma membrane. (B-B′′) The Cftrpd1048-GFP fusion protein has a reduction in plasma membrane localization. Arrows point to membrane localization, arrowheads denote the internal pool. (C) A western blot for GFP detects Cftr-GFP and Cftrpd1048-GFP expressed from HEK293 cells. Scale bars: 50 μm. |
|
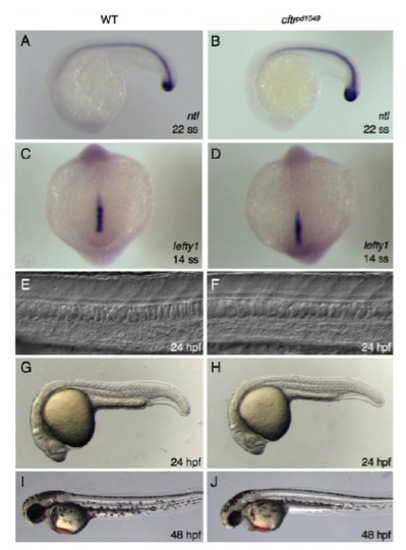
cftrpd1048 mutants do not have midline defects. (A,B) In situ hybridization of WT and mutant embryos for ntl. (C,D) In situ hybridization of WT and mutant embryos for lefty1. (E,F) DIC images of WT and mutant showing normal notochord and floorplate morphology. (G-J) Whole-mount brightfield images of WT and mutant embryos at (G,H) 24 hpf and (I,J) 48 hpf. |
|
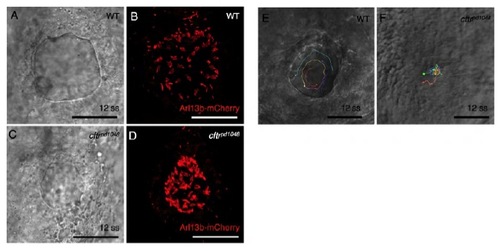
Motile cilia are crowded in cftrpd1048 mutants. (A,B) Brightfield and confocal imaging of 12 ss embryos injected with RNA encoding Arl13b-mCherry show normal morphology of KV, which contains a large number of motile cilia. (C,D) In the hypomorphic cftrpd1048 allele, reduced lumen size leads to crowding of motile cilia. (E,F) Tracking of fluorescent beads in WT and cftrpd1048KV. Scale bars: 50 μm. |