- Title
-
Skeletogenic fate of zebrafish cranial and trunk neural crest
- Authors
- Kague, E., Gallagher, M., Burke, S., Parsons, M., Franz-Odendaal, T., and Fisher, S.
- Source
- Full text @ PLoS One
|
Cre recombinase permanently activates reporter gene expression in sox10 expressing cells of the zebrafish embryo. A) Diagram of transgenes used to genetically mark neural crest descendants; Cre activity under control of the Sox10 enhancer results in excision of the floxed first coding sequence in each reporter. In the first, dsRed is excised, leading to persistent expression of egfp under control of the ubiquitous ef1a promoter. In the second, cyan fluorescent protein (cfp) excision leads to persistent expression of nuclear mCherry (nucCh). B–D) At 24 hours post fertilization, egfp expression resulting from Cre activation (B, C) shows the same pattern as the expression under direct control of the Sox10 enhancer (D). E, F) Expression of cre is shown by in situ hybridization of a Sox10:egfp transgenic embryo. E) Early expression of cre is seen to the anterior extent of NC (arrow) flanking the neural keel at 6 somites. F) Expression persists in the mandibular (m), hyoid (h), and branchial (b) clusters of NC at 14 somites. G) At 30 hours, doubly transgenic embryos show robust expression of egfp in cells known to be derived from neural crest, including in the branchial arches (br), and in the otic vesicle (ov). B-D, G are side views with anterior to the left; E and F are dorsal views with anterior to the left. EXPRESSION / LABELING:
|
|
GFP expression persists and reveals pattern of neural crest derivatives in head. A-K represent successive Z-stack projections of five confocal sections each, moving from ventral to dorsal through the head of a 10dpf doubly transgenic embryo. Images have been colored so that green represents GFP+ cells (NC derivatives) and magenta dsRed+ cells (non-NC). Note that throughout the remaining figures, the label associated with NC (GFP or nucCherry) is always shown as green in the two–color overlays. Cartilages known to be NC-derived, including Meckel′s cartilage (B), the ethmoid plate (F), and palatoquadrate (G) are labeled. Also GFP+ are cells in specific areas of ossification, including the dentary (A) and the anguloarticular (E) surrounding Meckel′s cartilage, and the maxilla and premaxilla (J) of the upper jaw. Note also the GFP+ nerve plexus in the lip taste buds (arrows in C), representing their innervation by NC-derived cells of the facial ganglia. Non-NC-derivatives, such as the intermandibularis anterior (ima) and interhyoideus (ih) muscle masses, remain dsRed+. Abbreviations for skeletal structures are listed in Table 3. EXPRESSION / LABELING:
|
|
Chondrocytes and osteoblasts of the pharyngeal skeleton are NC-derived. A-I) Transgenics carrying a reporter that activates nuclear-Cherry expression following Cre activation (A, D, G) were crossed to -1.4col1a1:egfp transgenics, in which all cartilage cells are GFP+ (B, E, H). At 4 dpf, cells within the ceratohyal (A-C), hyosymplectic (D-F) and Meckel′s (G-I) cartilages have nucCh+ nuclei, indicating they are NC-derived. The GFP cells surrounding the cartilages, largely representing perichondral cells or osteoblast precursors, are also NC-derived. J-L) The reporter transgene switches from dsRed to GFP expression following Cre activation. At 10 dpf (J, K), cartilage cells of the ceratohyal (J) and Meckel′s (K) cartilages are GFP+, as are the cells surrounding them, indicating that the bone replacing the cartilages is also NC-derived. Bones forming via membranous ossification, such as the opercle (L), are also NC-derived. |
|
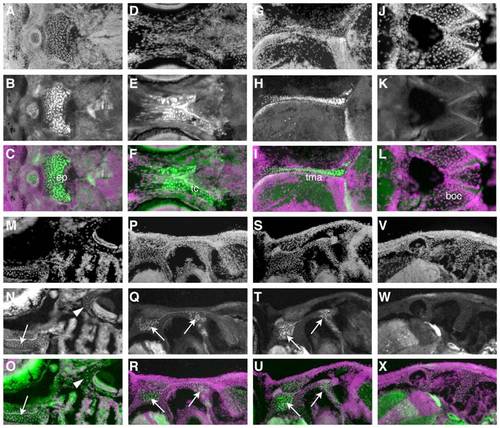
The chondrocranium is of mixed origin. A-R) Immunohistochemistry for GFP shows composition of cartilages with cellular resolution. In each set of three images, the first shows the DAPI counterstain (A, D, G, etc.), and the second (B, E, H, etc.) the GFP immunoreactivity. The third image in each group, the overlays, are pseudocolored with green representing GFP immunoreactivity and magenta the DAPI counterstain. The most anterior cartilages in the base of the skull, such as the ethmoid plate (A-C), trabeculae cranii (D-F), and taeniae marginalis anterior (G-I) are NC-derived. More posterior cartilages, like the basioccipital (J-L), contain no NC. M-O) A horizontal section at 14 dpf illustrates a more anterior NC-derived cartilage (arrow), the trabeculae cranii, and more posterior negative cartilage around the ear (arrowhead). P-X) Successive sections through a single fish at 44 dpf show that cartilage at intermediate locations, such as around the ear, is composed of a mix of NC (arrows) and non-NC cells in more ventral sections (P-R), and shows no NC-derived cells more dorsally (V-X). EXPRESSION / LABELING:
|
|
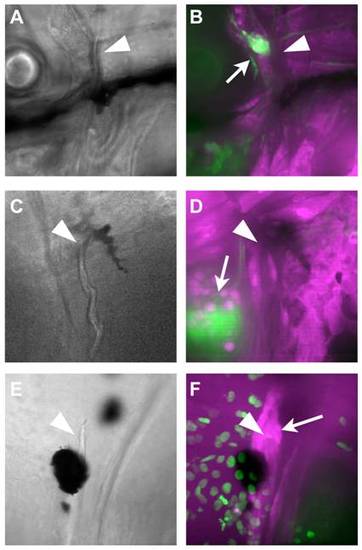
NC contribution to mineralized tissues of the adult skull. A) Bones derived by ossification of the pharyngeal arch cartilages are also NC-derived, seen in a horizontal section through a 44 dpf fish stained for GFP immunoreactivity. B) The odontoblasts of the pharyngeal teeth on the fifth ceratobranchial express the RUNX2:egfp transgene (B′) and are also NC-derived, seen by nucCh+ nuclei (B′′). C, D) The scleral cartilages are NC-derived, shown by GFP immunohistochemistry (C), as are the ossicles derived by their ossification, seen as GFP+ in freshly dissected tissue (D). E) The parasphenoid shows no NC contribution, as seen in freshly dissected tissue. The scattered nucCh+ cells (E′, E′′) are in associated soft tissue. The kinethmoid (F) also shows no NC contribution, although some of the associated soft tissue is NC-derived (nucCh+ in F′, F′′). G-G′′) Sections through the kinethmoid cartilage at 7 weeks show no GFP expression (G2) by immunohistochemistry (G is DAPI counterstain, G′′ shows overlay). H, I) Dissections were used to determine the status of unresolved bones throughout the skull, e.g. the pterosphenoid is nucCh+ in freshly dissected tissue (I) and the supratemporal is not. J, K) The anterior portion of the frontal bones are NC-derived, seen as GFP+ cells in a live, 6-week-old fish (J), and also as nucCh+ cells in freshly dissected tissue (K). Dotted lines indicate the location of the coronal suture. The more posterior portion of the frontal bones, and the other flat bones of the skull, show no evidence of NC contribution. L-M′′) Sections through the anterior frontal bone of a 7-week fish (L-L′′) show GFP+ osteoblasts by immunohistochemistry (arrowheads in L2) aligned under the acellular bone matrix (bracket in L), as well as GFP+ cartilage cells in the underlying epiphyseal bar. A similar section through the posterior frontal bone (M-M′′) shows no GFP expression in the osteoblasts (M′). In each set of panels, the first is the DAPI counterstain, the second the GFP immunohistochemistry, and the third the overlay. EXPRESSION / LABELING:
|
|
NC does not contribute to the cleithrum. A-D) At 10 dpf (A, B) and 16 dpf (C, D), Z-stack projections of confocal sections through the area surrounding the dorsal end of the cleithrum (arrowheads) reveal no GFP+ cells. The NC-derived glia of the lateral line ganglia are clearly visible in the same fields of view (arrows). E, F) To localize the osteoblasts, fish carrying the nucCh reporter were crossed with RUNX2:egfp transgenics, in which the osteoblasts are GFP+. The dorsal tip of the cleithrum (arrowhead in E, F) is surrounded by osteoblasts (arrow in F), which do not have nucCh+ nuclei, indicating they are not NC-derived. EXPRESSION / LABELING:
|
|
The scleroblasts of the caudal fin are NC derived. A) At 8 dpf, NC-derived cells (GFP+; arrow) can be seen clustered around the tip of the notochord (nc). B, C) By 16 dpf, there are more GFP+ cells; some are located more distally in the fin, although many are still close to the notochord. D-H) At 21 dpf, the caudal fin contains well-formed lepidotrichia (le in D), which are associated with GFP+ cells (E). F-H) To confirm the identity of the cells as osteoblasts, fish carrying the nucCh reporter were crossed with RUNX2:egfp transgenics, in which the osteoblasts are GFP+. The osteoblasts have nucCh+ nuclei, indicating they are NC-derived (G), and they are located both within (arrowhead) and immediately outside (arrow) the lepidotrichia (H). |