- Title
-
Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation
- Authors
- Hinits, Y., Pan, L., Walker, C., Dowd, J., Moens, C.B., and Hughes, S.M.
- Source
- Full text @ Dev. Biol.
|
Mef2ca and mef2cb expression during early cardiogenesis. In situ mRNA hybridisation of indicated genes, or immunodetection of Mef2ca/cb protein (G) for wild type (A–E, G–I) and Tg(myl7:EGFP) (F) in dorsal (A and B) or lateral (C–E) views of wholemount embryos or in flatmounts of dorsal views of the cardiac region, anterior to top (F–I). A and B. Mef2ca and mef2cb mRNAs accumulate in the bilateral heart fields in ALPM (black arrowheads) and adaxial cells (green arrowhead). C–E. Egr2b expression in rhombomeres 3 and 5, and ntl expression in the notochord positions the row of ventral cells in the ALPM (black arrowheads; D and E) that contain mef2ca and mef2cb, but not mef2aa mRNA (C). F. Confocal stack of Tg(myl7:EGFP) heart at 25 ss showing co-localisation of mef2cb mRNA (Fast Red) and EGFP. G. Mef2c protein in nuclei of a similar crescent of CMs spanning the midline. H and I. By 24 hpf, both mef2ca (H) and mef2cb (I) mRNAs are detected weakly in the heart tube, but mef2cb also accumulates strongly in the venous (blue arrow) and arterial (pink arrow) poles of the heart. Scale=100 μm (A–F), 20 μm (G–I). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) EXPRESSION / LABELING:
|
|
Early cardiomyocytes fail to differentiate after loss of Mef2c function. A–F. In situ mRNA hybridisation for myl7, vmhc and smyd1b in wild type control (A), mef2cab1086+mef2cb MO (B), mef2cab1086 (C) mef2cb MO (D), mef2cbfh288 (E) and mef2cab1086;mef2cbfh288 (F) embryos, shown in wholemounts in dorsal view, anterior to top. Loss of both mef2ca and mef2cb function greatly reduces myl7, vmhc and smyd1b mRNAs in the bilateral heart fields (arrowheads; A,B and F). Mef2cab1086 mutant embryos have weak myl7 and smyd1b mRNAs early, but recover later, and show no change in vmhc (C). Mef2cb single morphants or mef2cbfh288 mutant show no changes (D). G. Immunostaining for MyHC (A4.1025) in 24 ss mef2cab1086;mef2cbfh288 embryos and their siblings, shown in wholemounts in dorsal view, anterior to top (left panel) and lateral view, anterior to left (right panel). No MyHC is detected in the heart, whereas somitic muscle appears normal (white arrowheads). Scale=100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Redundant and specific functions of Mef2ca and Mef2cb drive cardiomyogenesis and heart tube formation. Immunodetection of MyHC (confocal stacks, top panels) or in situ mRNA hybridisation for indicated genes (A–F, lower panels and G-J) in hearts of 24 hpf zebrafish embryos shown in a dorsal view, anterior to top. A,B and F. Loss of both mef2ca (tn213 allele, in MyHC and b1086 allele, in bmp4 and myl7+vmhc) and mef2cb function (B and F) led to lack of all markers, compared with control (A). Note the few cells expressing myl7 only (black arrowhead) or both myl7 and vmhc (white arrowhead). C.Mef2ca mutants have a normal heart. D and E.Mef2cb morphants have a shortened heart with substantial loss of both atrial and ventricular volume, yet mef2cbfh288 mutants have a normal heart. G–I. Loss of Mef2c function with mef2d/c MO ablated all actin (acta1b, G), tnnc2 (H), mybpc1 (I) and nppa (J) mRNAs. Scale=100 μm. |
|
Loss of Mef2c function abolishes sarcomeric gene expression. Hearts of 48–50 hpf (A–F) or 72 hpf (G) mef2cab1086;mef2cbfh288 mutant embryos and their siblings in bright field (A), after immunodetection (C,D and G) or in situ mRNA hybridisation (B,E and F) shown in lateral view, anterior to left (A and G) or ventral (B–F) view, anterior to top. A.mef2cab1086;mef2cbfh288 embryos had a tiny residual heart (arrow) and cardiac chamber edema. B,E and F. Double mutant embryos lacked almost all myl7, vmhc, bmp4 and nppa mRNAs as well as MyHC and Mef2 proteins in ventricle (v), atrium (a) or AV canal (red arrows in F) except for one (arrows in C,E) or two small heart structures expressing these markers. C. Confocal stack showing genotyped double mutant expressing low levels MyHC and nuclear Mef2 in a residual myocardial tissue. Sibling presented is mef2ca+/b1086;mef2cb+/+. D. No MyHC or atrial MyHC is detected in mef2d/c morphants. G. Immunodetection for GFP (atrium and ventricle, green) and Elastin (bulbus arteriosus, ba) in hearts of three different genotyped mef2cab1086;mef2cbfh288;Tg(myl7:EGFP)twu26 showing variation in residual differentiated myocardial and bulbus tissue, compared to the normal heart of a mef2ca+/b1086;mef2cb+/fh288;Tg(myl7:EGFP)twu26 sibling. Scale=100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cardiomyocytes of mef2ca;mef2cb dual loss of function are specified but developmentally arrested. In situ mRNA hybridisation for indicated genes (wholemounts in dorsal view, anterior to top, except J, ventral view). A–C. ALPM expression (arrowheads) of nkx2.5 (A), hand2 (B) and gata4 (C) at 12 ss is unaffected by lack of Mef2ca and Mef2cb. Notochord and rhombomeres 3 and 5 are marked by ntl (blue) and egr2b (red), respectively. D,E.Tbx20 and gata6 mRNAs are present but disorganised in the heart region of mef2d/c morphants and mef2cab1086;mef2cbfh288 mutant embryos (lacking myl7 expression in red, E, right panel) compared to the typical ring-shape in control and sibling embryos. F. During heart cone stage (22 ss), nkx2.5 mRNA is abolished. G. hand2 mRNA is enhanced in a sheet of cells spanning the cardiac region but not elsewhere. Pharyngeal pouch expression is unchanged (white arrowheads). H, Whereas myocardial cells are undifferentiated in mef2d/c morphants and mef2cab1086;mef2cbfh288 mutants (myl7, red), the endocardium is expanded and cdh5 is up-regulated. I. Endocardium (cdh5, arrow) lines the myocardium (myl7, red) in the normal looped heart of a sibling embryo. In mef2ca;mef2cb mutant embryos, little endocardial makrer is co-localised with the residual myocardium (arrows). Extra cdh5-expressing tissue is detected in the cardiac region (arrowheads). Scale=100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
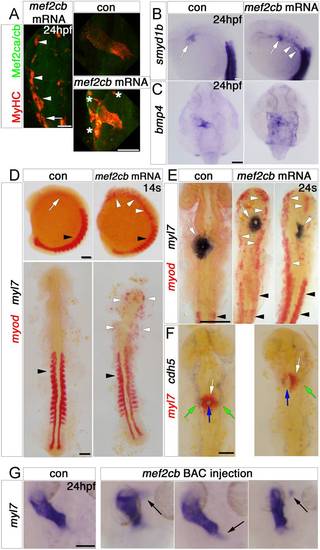
Mef2cb overexpression induces skeletal muscle at the expense of myocardial and endothelial cells. In situ mRNA hybridisation (or immunodetection, A) for indicated genes, shown as wholemounts in dorsal (G and C) or lateral (B and D, top panel) views, or as flatmounts in dorsal view (A and D, bottom panel, E and F). A. Injection of mef2cb RNA results in many ectopic muscle cells expressing strong Mef2ca/cb and MyHC at 24 hpf (white arrowheads) anterior to the first somite (white arrow). In the heart, MyHC and Mef2ca/cb are mosaically stronger than in control embryos. Asterisks=ectopic muscle. B.Smyd1b is upregulated in CMs (white arrow), and in ectopic muscle in the head region (white arrowheads). C. Expression of bmp4 is upregulated in much of a sheet of cardiogenic cells but not elsewhere in the embryo. D. In 14 ss control embryos, myod mRNA is expressed in somites (black arrowheads) and myl7 is expressed weakly in CMs (white arrow). Embryos injected with mef2cb RNA express no ectopic myl7, but have ectopic myod mRNA in the head region (white arrowheads). E. By 24 ss, high levels of ectopic myod correlated with reduction or lack of myl7-expressing CMs (right panel) and defective brain development. F. At 24 ss, mef2cb RNA-injected embryos that had fewer myl7-expressing CMs (white arrows) also had less cdh5 expression in vascular endothelium (green arrows) and disorganised endocardium (blue arrows). G. Compared to wild type control (leftmost panel), three examples of embryos injected with mef2cb BAC DNA show ectopic myl7 mRNA either contiguous with (left panels) or detached from (right panel) the arterial (flanking panels) or venous (middle panel) poles. Scale=100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
|
Loss of mef2cb function results in no heart phenotype. In situ mRNA hybridisation for myl7, vmhc and bmp4 (A,B, ventral view, anterior to top) and immunodetection of DM-GRASP (zn5) and Elastin (C, lateral view, anterior to left) of hearts at indicated stage of genotyped mef2cbfh288mutants and their siblings. A,B. mef2cbfh288 mutants had a normal looped heart and normal expression of chamber markers, and bmp4 mRNA in OFT (blue arrow), IFT (purple arrow) and AV canal (red arrows). C. Confocal stacks of genotyped mef2cbfh288 mutants and siblings with a normal looped heart with developed chambers and bulbus arteriosus. Scale=100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
Mef2cb mRNA and protein are upregulated after mef2cb over-expression. EXPRESSION / LABELING:
|
|
Mef2c protein is downregulated in mef2ca mutants. EXPRESSION / LABELING:
|
|
Mef2cb morpholinos are specific and efficient in targeting translation and splicing of mef2cb. |
|
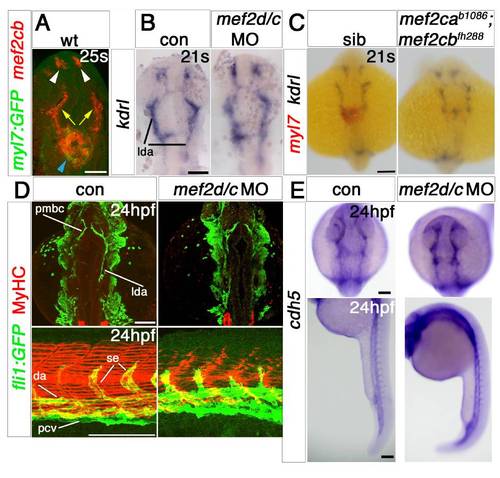
Endothelial markers are little affected by Mef2 knockdown. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Mef2ca and Mef2cb dual loss of function abolishes myocardial differentiation. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Mef2ca and Mef2cb dual loss of function abolishes myocardial differentiation. |
|
Mef2ca and Mef2cb are expressed in hand2 knockdown embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Mef2ca mutant has a normal heart. EXPRESSION / LABELING:
|
|
Mef2cb morphants have a linear defective heart and loss of bulbus arteriosus. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 369(2), Hinits, Y., Pan, L., Walker, C., Dowd, J., Moens, C.B., and Hughes, S.M., Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation, 199-210, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.