- Title
-
Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo
- Authors
- Wang, X., Ono, Y., Tan, S.C., Chai, R.J., Parkin, C., and Ingham, P.W.
- Source
- Full text @ Development
|
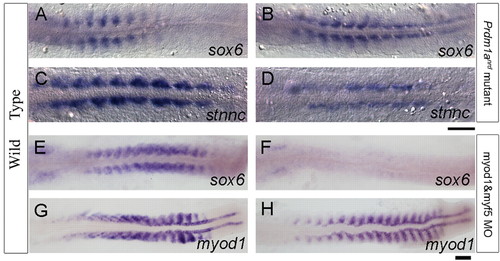
Negative and positive regulation of sox6 in the zebrafish myotome. (A,B) sox6 expression is restricted to the fast muscle domain in a wild-type 10-somite stage embryo (A) but is ectopically expressed in adaxial cells in a prdm1anrd mutant at the same stage. (C,D) This derepression correlates with downregulation of stnnc expression in adaxial cells in the prdm1anrd mutant (D) relative to wild type (C). (E,F) sox6 transcript persists in the fast-twitch muscle domain and also accumulates in the otic placode in the wild type at the 18-somite stage (E), whereas the myod1;myf5 morphant shows loss of sox6 expression specifically from the muscle progenitors. (G,H) Expression of myod1 in wild-type (G) and myod1;myf5 morphant (H) embryos. Scale bars: 50 μm. EXPRESSION / LABELING:
|
|
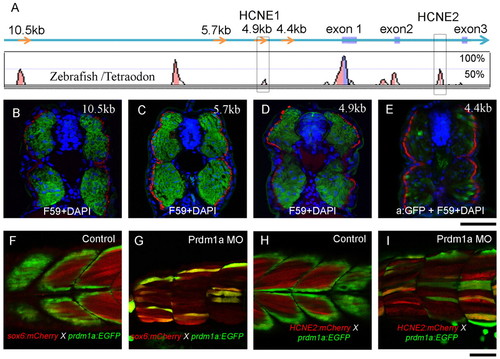
Conserved cis-elements recapitulate the sox6 expression pattern in the myotome. (A) Conserved non-coding elements (represented by peaks) upstream and downstream of the transcription start site revealed by alignment of the zebrafish and Tetraodon sox6 genomic sequences. Several of these are also highly conserved in mammals (see Fig. S3 in the supplementary material), including HCNE1 and HCNE2 (red boxes). (B-E) Transverse sections through mid-trunk regions of 48 hpf embryos transgenic for sox6 reporter genes; slow-twitch fibres are visualised with mAb F59 (red), nuclei by DAPI staining (blue). The 10.5 kb, 5.7 kb and 4.9 kb fragments drive bright and specific EGFP (green) expression exclusively in fast-twitch fibres; by contrast, the 4.4 kb fragment drives low-level mosaic expression of EGFP, detectable only in fixed specimens using anti-EGFP antibody (E). (F-I) Optical sagittal sections of 48 hpf Tg(PACprdm1:GFP)i106 embryos carrying the sox6 5.7 kb or HCNE2 mCherry reporter constructs. Both constructs are ectopically expressed (red) in fibres derived from adaxial cells (green) following injection of Prdm1a morpholino (G,I). Scale bars: 50 μm. EXPRESSION / LABELING:
|
|
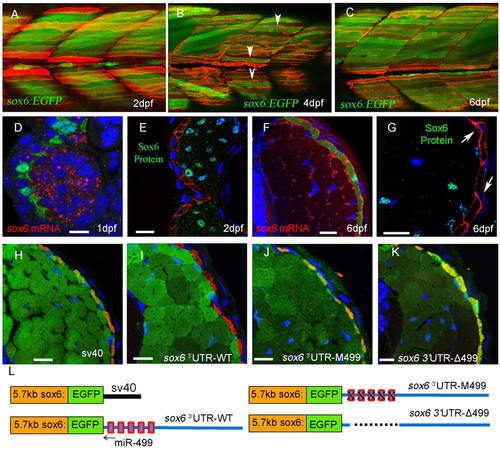
Derepression of sox6 transcription but not translation in slow-twitch fibres. (A-C) Optical sagittal sections of posterior trunk region of Tg(5.7 kb sox6:EGFP)i253; Tg(9.7 kb smyhc1:lyn-tdTomato)i261 zebrafish larvae. At 2 dpf (A), EGFP expression is restricted exclusively to fast-twitch fibres (green) lying beneath the superficial slow-twitch fibres (red). At 4 dpf (B), EGFP expression is detectable in a few slow-twitch fibres (arrowheads). By 6 dpf (C), all slow-twitch fibres express EGFP. (D-G) Transverse cryostat sections of Tg(BACsmyhc1:GFP)i108 (D,F) or Tg(9.7 kb smyhc1:lyn-tdTomato)i261 (E,G) embryos/larvae hybridised with a probe for sox6 mRNA (red) or stained with anti-Sox6 antibody (green), respectively. Arrows indicate slow-twitch fibre nuclei. (H-L) Transverse cryostat sections of 6 dpf larvae carrying the 5.7 kb sox6:EGFP reporter constructs with differing 3′UTRs, as illustrated in L. (H) Expression of EGFP driven by the 5.7 kb reporter construct with the SV40 3′UTR in slow-twitch fibres, as identified by mABF59 staining (red), is repressed by replacement of the SV40 3′UTR with the endogenous sox6 3′UTR (I). Mutation (J) or deletion (K) of the five putative miR-499 target sites restores ectopic EGFP expression in slow-twitch fibres. Note the increased levels of EGFP expression caused by deletion of the miR-499 sites (K). Scale bars: 10 µm. |
|
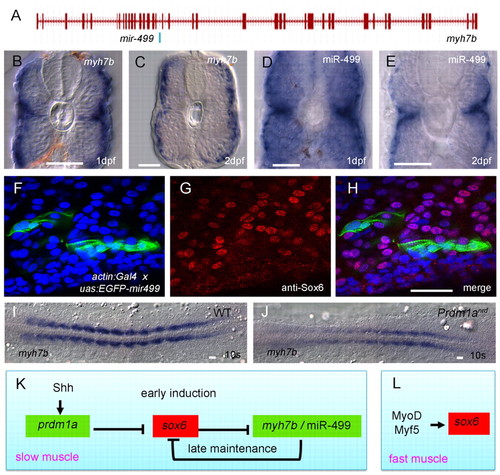
Prdm1a promotes the spatially restricted expression of myh7b and mir499, which is sufficient to repress sox6 translation. (A) Exon-intron structure of the myh7b gene showing the location of mir499 in intron 18. (B-E) Transverse sections of wild-type embryos hybridised with probes for myh7b or miR-499 transcripts, both of which are restricted to the superficially located slow-twitch fibres and the medially located muscle pioneers, a subset of the slow-twitch lineage. (F-H) Sagittal optical sections of a 24 hpf Tg(actin:GAL4) embryo injected with an EGFP-mir499 construct under UAS control; fibres expressing the construct (F, green) show significant reduction in nuclear accumulation of Sox6 protein (G, red) as revealed by the merged image (H); nuclei are stained with DAPI (blue). (I,J) Flat mounts of 10-somite stage wild-type and prdm1anrd mutant embryos hybridised with probe for myh7b, showing strong downregulation in the absence of Prdm1a function. (K,L) Gene regulatory network underlying the specification and maintenance of slow-twitch and fast-twitch muscle lineages in zebrafish. Scale bars: 20 μm. EXPRESSION / LABELING:
|
|
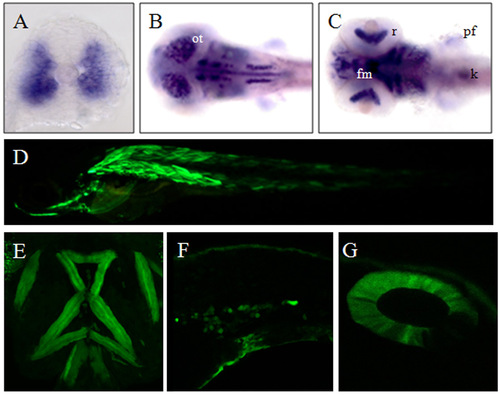
Expression pattern of sox6 and the 10.5kb sox6:EGFP transgene. (A) Transverse section of an 18-somite stage embryo showing sox6 mRNA accumulation in the fast muscle domain. (B) Dorsal view of a 3 dpf larva showing accumulation of sox6 transcript in the optic tectum (ot) and other parts of the brain. (C) Ventral view of the same larva showing sox6 transcript in the facial muscle (fm), retina (r), pectoral fin (pf) and kidney (k). (D) Transient expression of Tg(10.5kb sox6:EGFP) in the skeletal muscle after co-injection with tol2 mRNA. (E-G) Expression of Tg(10.5 kb sox6:EGFP) in the ventral head muscles, intermediate cell mass (F) and otic vesicle (G) of stable transgenic larvae. |
|
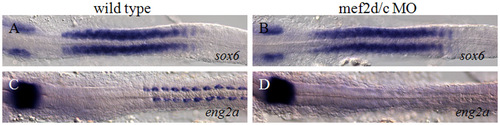
Myocyte-specific enhancer factor 2 (Mef2) is not required for sox6 expression. (A,B) Accumulation of sox6 mRNA at the 18-somite stage in a wild-type embryo (A) and a mef2c/d morphant embryo (B). (C,D) Accumulation of eng2a transcript in a wild-type embryo (C) and in a mef2c/d morphant embryo (D). Note the loss of transcript specifically from the myotome of the morphant. |