- Title
-
The PAF1 complex differentially regulates cardiomyocyte specification
- Authors
- Langenbacher, A.D., Nguyen, C.T., Cavanaugh, A.M., Huang, J., Lu, F., and Chen, J.N.
- Source
- Full text @ Dev. Biol.
|
Fig. 1. Mutation in zebrafish ctr9 perturbs primitive heart tube elongation and causes a reduction in cardiomyocyte cell number. (A and B) Lateral views of 24 hpf wild type (A) and LA961 mutant (B) embryos. LA961 mutants can be distinguished from wild type embryos based on missing otoliths (arrowhead) and pericardial edema (arrow). (C and D) Wild type and LA961 mutant embryos analyzed for cmlc2 expression at 24 hpf. Wild type embryos have an elongated primitive heart tube (C), whereas LA961 mutants have a clump of cardiomyocytes at the midline (D). (E) Graph of number of GFP-positive cardiomyocytes in wild type (WT) and LA961 mutant embryos. Error bars indicate standard deviations. (F) Lateral view of wild type (top) and LA961 mutant (bottom) embryos at 35 hpf. (G) Diagram of LA961 mapping. Estimated genetic distance is in centimorgans (cM). (H) Diagram of Ctr9WT and Ctr9LA961 proteins. Grey regions are predicted tetratricopeptide repeat domains and black regions are predicted nuclear localization domains. |
|
Fig. 2. Zebrafish PAF1C components localize to the nucleus and interact with each other in vivo. (A–A3) Panel (A3) shows the colocalization of Myc-tagged Ctr9 protein (red, A) and nuclear DAPI staining (blue, A2). (B) Flag-tagged Cdc73 (red) and DAPI staining (blue) colocalize in the nucleus. (C) Flag-tagged Paf1 (red) and DAPI staining (blue) colocalize in the nucleus. (D) Flag-tagged Rtf1 (red) and DAPI staining (blue) colocalize in the nucleus. (E) Flag-Paf1, Flag-Cdc73, and Flag-Leo1 coimmunoprecipitate with Myc-Ctr9. Input row shows 1 embryo equivalent of total protein lysate. IP row shows immunoprecipitated protein from 20 embryo equivalents of total protein lysate. |
|
Fig. 3. Loss of Ctr9, Cdc73, Paf1, and Rtf1 causes defects in myocardial migration, cardiomyocyte cell number, and primitive heart tube elongation. (A–E) Dorsal views of cmlc2 expression in 24 hpf wild type and morphant embryos. Wild type embryos have an elongated primitive heart tube at 24 hpf (A), whereas ctr9 (B), cdc73 (C), and paf1 (D) morphants have a small clump of cardiomyocytes situated at the midline. Rtf1 morphants have no detectable cmlc2 expression at 24 hpf (E). (F–O) Dorsal views of nkx2.5 expression in 16S (F–J) and 20S (K–O) stage wild type and morphant embryos. In wild type embryos, myocardial progenitors arise in the LPM, migrate towards the midline, and fuse to form a cardiac cone (F and K). In ctr9 morphants (G and L), nkx2.5 expression is reduced and myocardial migration and fusion are delayed. Cdc73 morphants (H and M) also display delays in the migration and fusion of the myocardial progenitors, but have relatively normal nkx2.5 expression levels. Paf1 morphants (I and N) and rtf1 morphants (J and O), like ctr9 morphants, display reduced nkx2.5 expression and delayed migration and fusion of the myocardial progenitors. Expression of nkx2.5 in rtf1 morphants is barely detectable. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fig. 4. Ctr9, Cdc73, and Paf1 are required for AV boundary specification. (A–D) Cardiac expression of vmhc in 48 hpf embryos. In wild type (A), ctr9LA961 mutant (B), cdc73 morphant (C), and paf1 morphant (D) embryos, vmhc expression is detected primarily in the ventricle. (E–H) Cardiac expression of amhc in 48 hpf embryos. In wild type (E), ctr9LA961 mutant (F), cdc73 morphant (G), and paf1 morphant (H) embryos, amhc expression is detected primarily in the atrium. (I–L) Cardiac expression of bmp4 in 48 hpf embryos. In wild type embryos (I), bmp4 expression is detected in the myocardium of the ventricle, sinus venosus, and AV boundary (arrow). Ctr9LA961 mutant (J), cdc73 morphant (K), and paf1 morphant (L) embryos display ventricular expression of bmp4, but have no discernable AV boundary expression. (M–P) Cardiac expression of tbx2b in 48 hpf embryos. In wild type embryos, tbx2b is expressed by the myocardium of the AV boundary (arrow, M). Ctr9LA961 mutant (N), cdc73 morphant (O), and paf1 morphant (P) embryos instead aberrantly express tbx2b throughout the heart. (Q–T) Cardiac expression of notch1b in 48 hpf embryos. Notch1b is expressed by the AV boundary endocardium in wild type embryos (arrow, Q), but is not expressed in the hearts of ctr9LA961 mutants (R), cdc73 morphants (S), and paf1 morphants (T). |
|
Fig. 5. The PAF1C is required for the specification of cardiomyocytes from the anterior lateral plate mesoderm. (A–L) Dorsal views of hand2 (A–C), gata4 (D–F), gata5 (G–I), and tbx20 (J–L) expression in 16S stage wild type and morphant embryos. In wild type embryos, expression of hand2 (A), gata4 (D), and gata5 (G) is detected in the anterior lateral plate mesoderm. In ctr9 and rtf1 morphants, hand2 (B and C), gata4 (E and F), and gata5 (H and I) are expressed in a domain similar to that of wild type embryos. (J–L) In wild type embryos, tbx20 is expressed by cardiomyocyte progenitors in the ALPM (J). Expression of tbx20 is reduced in ctr9 morphants (K) and strongly reduced in rtf1 morphants (L) at the 16S stage. EXPRESSION / LABELING:
PHENOTYPE:
|
|
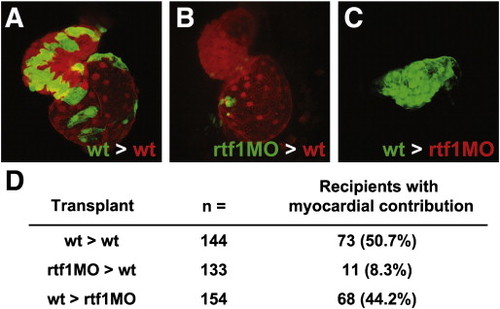
Fig. 6. Rtf1 regulates cardiomyocyte specification cell autonomously. (A–C) Confocal projections showing a ventral view of the heart in two day-old transplanted embryos. Wild type donor cells contribute robustly to the heart when transplanted into wild type recipients (A), while rtf1 morphant cells exhibit limited contribution when transplanted into a wild type recipient (B). Wild type cells are also able to generate myocardial tissue when transplanted into an rtf1 morphant host. (D) Summary of the results from transplantation experiments showing the contribution of donor cells to the myocardium of recipient embryos. |
|
LA961 is a mutation in ctr9. (A–D) In contrast to wild type embryos (A), LA961 mutant embryos at 2 dpf (B) exhibit pericardial edema, a lack of pigmentation, and a curved body axis. Injection of 200 pg of ctr9WT mRNA rescues the cardiac, pigment, and body axis defects of LA961 mutants (C). Injection of 0.75 ng of ctr9MO phenocopies the LA961 mutant phenotype (D). PHENOTYPE:
|
|
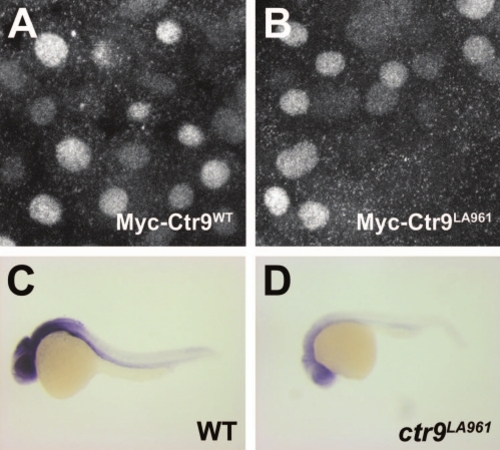
Ctr9LA961 transcripts undergo nonsense-mediated decay. (A and B) Myc-Ctr9WT (A) and Myc-Ctr9LA961 (B) both localize to the nucleus. (C and D) Lateral views of ctr9 expression in 1 day post fertilization wild type and ctr9LA961 mutant embryos. Expression of ctr9 is very reduced in ctr9LA961 mutants (D) compared to wild type embryos (C). EXPRESSION / LABELING:
|
|
Morpholinos targeting PAF1C component genes reduce target protein levels. (A–D) Western blots detecting PAF1C component proteins and β-actin protein levels in uninjected control and morpholino injected embryo lysates. (A) Ctr9 protein is reduced to 38% of control levels in ctr9MO injected embryos. (B) Paf1 protein is reduced to 43% of control levels in paf1MO injected embryos. (C) Cdc73 protein is reduced to 39% of control levels in cdc73MO injected embryos. (D) Rtf1 protein is reduced to 7% of control levels in rtf1MO injected embryos. EXPRESSION / LABELING:
|
|
Specification of cardiac progenitor cells is sensitive to Rtf1 dosage. (A–E) Dorsal views of cmlc2 expression in 24 hpf wild type and morphant embryos. Wild type embryos have an elongated primitive heart tube at 24 hpf (A), whereas embryos injected with 0.1 ng, 0.25 ng, and 1.5 ng of rtf1MO have no detectable cardiac tissue (B–D). Embryos coinjected with ctr9MO, cdc73MO, and paf1MO (3KD) have a small clump of cardiomyocytes situated at the midline (E). (F–J) Dorsal views of nkx2.5 expression in 16S stage wild type and morphant embryos. In wild type embryos, cardiac progenitors have been specified in the ALPM (F). Injection of rtf1MO produces a dosage-dependent effect on nkx2.5 expression. Embryos injected with 0.1 ng rtf1MO (G) have a reduction in nkx2.5 expression compared to wild type. Expression of nkx2.5 is further reduced in embryos injected with 0.25 ng rtf1MO (H), and absent in embryos injected with 1.5 ng rtf1MO (I). Simultaneous knockdown of ctr9, cdc73, and paf1 does not result in a phenotype as severe as that seen in rtf1 morphants. (K–O) Dorsal views of cmlc2 expression in 16S stage wild type and morphant embryos. Mature cardiomyocytes that express cmlc2 are beginning to be specified as early as the 16S stage in wild type embryos (K), but are not present in 0.1 ng, 0.25 ng, and 1.5 ng, rtf1MO and 3KD morphant embryos at this stage (L–O). |
Reprinted from Developmental Biology, 353(1), Langenbacher, A.D., Nguyen, C.T., Cavanaugh, A.M., Huang, J., Lu, F., and Chen, J.N., The PAF1 complex differentially regulates cardiomyocyte specification, 19-28, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.