- Title
-
Rumba and Haus3 are essential factors for the maintenance of hematopoietic stem/progenitor cells during zebrafish hematopoiesis
- Authors
- Du, L., Xu, J., Li, X., Ma, N., Liu, Y., Peng, J., Osato, M., Zhang, W., and Wen, Z.
- Source
- Full text @ Development
|
rumbahkz1 and sambahkz2 are defective in definitive hematopoiesis. (A-B′) WISH of rag1 expression in 6 dpf wild-type (WT) zebrafish siblings (A,A′) and rumbahkz1-/- (B) and sambahkz2-/- mutants (B′). The red circles indicate the position of the thymus. (C-F′) WISH of αe1-globin (C-D′) and mpo (E-F′) in 22 hpf wild-type siblings (C,C′,E,E′) and rumbahkz1-/- (D,F), and sambahkz2-/- mutants (D′,F′). (G-J′) WISH of αe1-globin (G-H′) and mpo (I-J, I′-J′) in 6 dpf wild-type siblings (G,G′,I,I′) and rumbahkz1-/- (H,J) and sambahkz2-/- mutants (H′,J′). Insets in G-J′ show enlarged details of the CHT region. The blue arrowheads indicate the position of the kidney. EXPRESSION / LABELING:
PHENOTYPE:
|
|
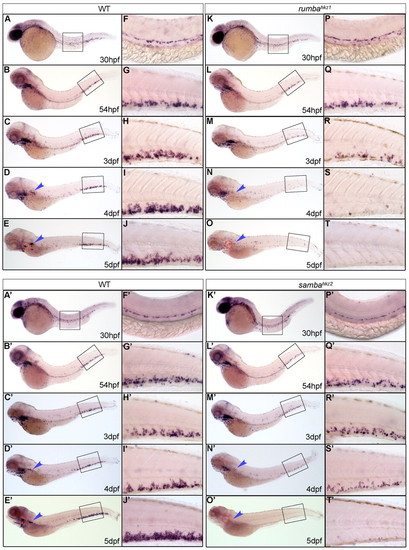
Definitive HSPCs are not sustained in the CHT of rumbahkz1 and sambahkz2 mutants. (A-T&strong;) WISH of c-myb in 30 hpf, 54 hpf, 3 dpf, 4 dpf and 5 dpf wild-type (WT) zebrafish siblings (A-E&strong;) and rumbahkz1-/- (K-O), and sambahkz2-/- mutants (K′-O′). The right-hand columns (F-J′,P-T′) represent the 20x magnification of the boxed regions in the left-hand columns (A-E′,K-O′). The red circles and the blue arrowheads show the position of the thymus and kidney, respectively. |
|
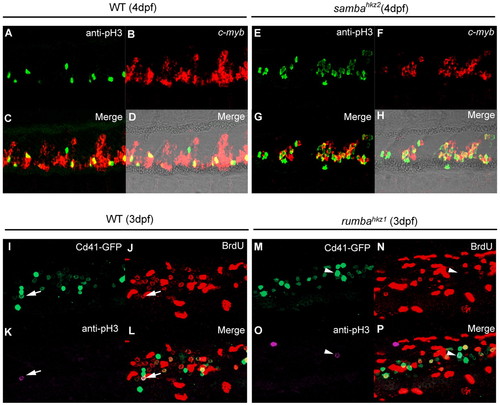
rumbahkz1 and sambahkz2 mutations cause different cellular defects in definitive hematopoietic cells. (A-H) Double staining of cmyb RNA and phospho histone 3 (pH3) protein in the CHT of 4 dpf wild-type (WT) zebrafish siblings (A-D) and sambahkz2-/- mutants (E-H). D and H show bright-field images with fluorescent staining overlaid. (I-P) Triple staining of Cd41-GFP, BrdU and phospho histone 3 (pH3) of 3 dpf wild-type siblings (I-L) and rumbahkz1-/- mutants (M-P). Arrows indicate Cd41-GFP, BrdU and pH3 triple-positive cell and arrowheads indicate Cd41-GFP and pH3 double-positive but BrdU-negative cell. EXPRESSION / LABELING:
|
|
rumba encodes a novel zinc finger protein. (A) The zebrafish rumba gene is mapped to a 184 kb region between two SSLP markers O82105 (17 recombinants in 1717 rumbahkz1-/- embryos) and L25805 (1 recombinant in 1717 rumbahkz1-/- embryos) in the LG16. This 184 kb region, covered by BAC zC287M15, zC106k4, zC239E16 and zK67N17, contains four predicted genes: the ETS domain transcriptional repressor PE1, a novel HMG protein, GSK-3α and the novel zinc finger protein. (B) The rumba gene consists of seven exons and a C-to-T substitution occurs in exon 6 in rumbahkz1 mutants. (C-E) WISH shows cmyb expression in the CHT of a 5 dpf a wild-type embryo (C), a rumbahkz1-/- mutant embryo (D) and a rumbahkz1-/- mutant embryo injected with synthesized wild-type rumba RNA (E). This phenotype is representative of 21 out of 33 mutant embryos observed, as indicated. (F) Protein sequence analysis predicts that Rumba consists of 16 C2H2 zinc-finger domains. (G-I) DAPI staining (G) and immunostaining with anti-Rumba antibody (H) reveals that Rumba is located in the nucleus as indicated by the colocalization of Rumba and DAPI (I). EXPRESSION / LABELING:
|
|
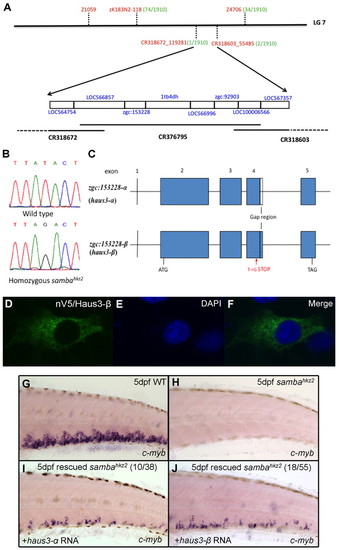
The sambahkz2 mutation is mapped to the haus3 gene in linkage group 7 of the zebrafish genome. (A) Positional cloning information. The sambahkz2 mutation is mapped to a three-overlapping-BAC region between markers CR318672_119281 and CR318603_55485 in LG7. This region contains eight candidate genes (blue). The simple sequence length polymorphism markers (SSLP) or single nucleotide polymorphism (SNP) markers used in mapping are labeled in red with their respective recombinants number (out of a total of 1910 homozygous sambahkz2 embryos) in green. (B) Sequencing results showing a nonsense mutation (a T-to-G transition) in haus3 (zgc:153228) gene. (C) Gene structure of haus3 (zgc:153228). Gap region indicates the absence of 48 bp in haus3-α (zgc:153228-α). (D-F) Immunohistochemical staining with the anti-V5 monoclonal antibody on pcDNA3.1/nV5-haus3-β-transfected cos-7 cells and DAPI staining of the same transfected cells. (G-J) In vitro synthesized full-length haus3 RNA rescues the mutant phenotype. WISH of cmyb expression in wild type (G), uninjected sambahkz2-/- mutant control (H) and sambahkz2-/- mutant embryos injected with either wild-type haus3-α (I) or haus3-β RNA (J). The phenotype seen in I and J is representative of 10 out of 38 and 18 out of 55 mutant embryos observed, as indicated. EXPRESSION / LABELING:
|
|
Rumba and Haus3 are cell autonomously required for maintaining HSPCs in the CHT. (A-H′) Confocal images of transplantation results in wild-type (WT) zebrafish sibling hosts (A-D′), rumbahkz1-/- mutant hosts (E-H), and sambahkz2-/- mutant hosts (E′-H′). Anti-GFP staining (A,A′,E,E′) and anti-αE1-globin staining (B,B′,F,F′) in the host CHT region are merged in C, C′, G and G′, as indicated and superimposed with their respective bright field views in D, D′, H and H′. Dotted lines indicate the position of the caudal vein. |
|
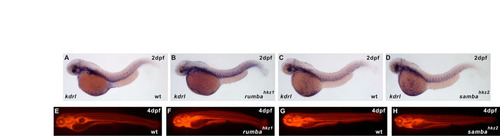
rumbahkz1 and sambahkz2 mutants have an intact vascular system. (A-D) WISH of kdrl (also known as flk1) expression in 2 dpf wild-type siblings (A,C) and rumbahkz1 (B) and sambahkz2 mutants (D). (E-H) Tetramethylrhodamine dextran marks the vessels in 4 dpf wild-type siblings (E,G) and rumbahkz1 (F) and sambahkz2 mutant (H). |
|
HSPCs are reduced in rumbahkz1-/-/Tg(cd41:eGFP) and sambahkz2-/-/Tg(cd41:eGFP) mutants. (A) 5 dpf wild-type Tg(cd41:eGFP) transgenic fish has a large number of GFP+ cells in the CHT region. (B) Superimposed image of panel A and its bright field view. (C) 5 dpf rumbahkz1-/-/Tg(cd41:eGFP) has very few GFP+ cells in the CHT region. (D) Superimposed image of panel C and its bright field view. (E) 5 dpf sambahkz2-/-/Tg(cd41:eGFP) has few GFP+ cells in the CHT. (F) Superimposed image of panel E and its bright field view. Dotted lines indicate the position of the caudal vein. |
|
Cell death assay of rumbahkz1 and sambahkz2 mutant embryos. (A-X) TUNEL and Cd41-GFP double staining of 3 dpf, 4 dpf and 5 dpf rumbahkz1 mutants (E-H,M-P,U-X) and wild-type siblings in the CHT (A-D,I-L,Q-T). (A′-X′) TUNEL and Cd41-GFP double staining of 3 dpf, 4 dpf and 5 dpf sambahkz2 mutants (E′-H′,M′-P′,U′-X′) and siblings in the CHT (A′-D′,I′-L′,Q′-T′). Arrows indicate GFP and TUNEL double positive cells. Arrowheads indicate GFP negative but TUNEL positive cells. (Y) Quantification of TUNEL and Cd41-GFP double-positive cells in the CHT of rumbahkz1 mutants and siblings. (Y′) Quantification of TUNEL and Cd41-GFP double-positive cells in the CHT of sambahkz2 mutants and siblings. Error bars indicate s.d. |
|
In vitro transcribed cRNA is stably expressed in injected embryos. RT-PCR was performed on 2-4 dpf embryos to examine the stability of injected rumba and haus3 RNA. SV40 polyA sequence was used to distinguish injected RNA from their endogenous RNA. elf1a expression was used as the control. |
|
rumba and haus3 genes are expressed ubiquitously during embryonic development. (A-E) WISH of rumba expression at the one-cell stage (A) and in 1 dpf (B), 2 dpf (C), 3 dpf (D) and 4 dpf (E) wild-type embryos. (A′-E′) WISH of haus3 (haus3-β) expression at the one-cell stage (A′) and in 1 dpf (B′), 2 dpf (C′), 3 dpf (D′) and 4 dpf (E′) wild-type embryos. Embryos in A and A′ are orientated with animal pole on top whereas embryos in B-E and B′-E′ are orientated with anterior to the left. |