- Title
-
Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4
- Authors
- Labalette, C., Bouchoucha, Y.X., Wassef, M.A., Gongal, P.A., Le Men, J., Becker, T., Gilardi-Hebenstreit, P., and Charnay, P.
- Source
- Full text @ Development
|
Spry gene expression in the early zebrafish hindbrain. (A-L) In situ hybridisations were performed with spry1 (A,D,G,J), spry2 (B,E,H,K) and spry4 (C,F,I,L) probes (blue) at the indicated somite (s) or epiboly (%) stages, shown as lateral views with anterior to the left (A-C,G-I) and flat-mounts with anterior at the top (D-F) or left (inset in I and J-L). Where indicated (D-F,J-L), double in situ hybridisation was performed with a krox20 probe (red) to allow precise localisation of r3 and r5. The inset in F shows the spry4 pattern without krox20 labelling. hb, hindbrain; tb, tailbud; tl, telencephalon; MHB, midbrain-hindbrain boundary. EXPRESSION / LABELING:
|
|
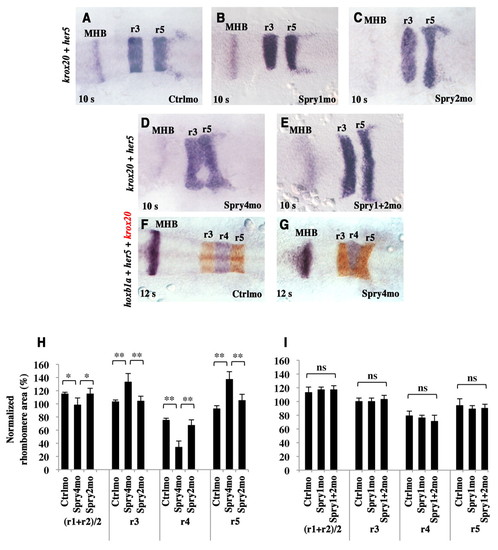
Spry4 loss-of-function results in hindbrain patterning defects. (A-G) Zebrafish embryos injected with either control morpholino (Ctrlmo) (A,F), Spry1mo (B), Spry2mo (C), Spry4mo (D,G) or both Spry1mo and Spry2mo (E) were collected at the 10-somite (A-E) or 12-somite (F,G) stage and subjected to in situ hybridisation for krox20 and her5, a marker of the MHB (A-E, both purple), or for krox20 (red), her5 and hoxb1a (purple) (F,G). Embryos are flat-mounted with anterior to the left. (H,I) Quantitative evaluation of rhombomere areas. Normalised areas were obtained by dividing each rhombomere area by one fifth of the area of the neural plate from r1 to r5. ns, not significant (P>0.05); *, P<0.004; **, P<0.0001; Student′s t-test. Errors bars indicate s.e.m. EXPRESSION / LABELING:
PHENOTYPE:
|
|
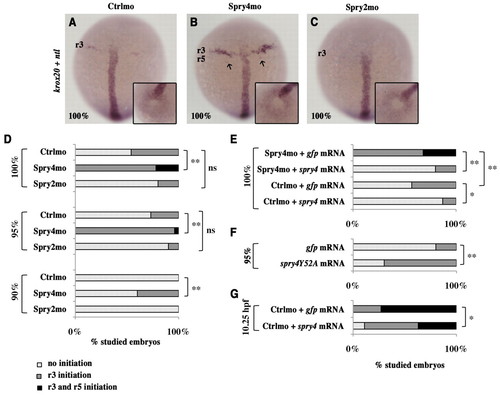
Spry4 controls the onset of krox20 expression. (A-C) Zebrafish embryos injected with either control morpholino (Ctrlmo) (A), Spry4mo (B) or Spry2mo (C) were collected at 100% epiboly and subjected to in situ hybridisation with krox20 and ntl probes (purple). Arrows in B indicate krox20 expression in a few r5 cells. The insets show tailbud views of the embryos, allowing determination of the developmental stage by evaluation of the closure of the tailbud, as revealed by ntl expression. (D-G) Distribution of embryos showing either no krox20 expression, limited expression in r3 or expression in both r3 and r5 at 90, 95 or 100% epiboly or at 10.25 hours post-fertilisation (hpf). ns, not significant; *, P<0.05; **, P<0.0001; χ2-test. |
|
FGF signalling is required for the appropriate onset of krox20 expression. (A-D,F-I) Zebrafish embryos were incubated in either DMSO carrier or SU5402 from 50% epiboly to 10.25 hpf (A,B) or from 50% epiboly to the 1-somite stage (10.5 hpf) (C,D) or from 80% epiboly to the 4-somite stage (F-I) and analysed by in situ hybridisation for krox20 and ntl (A-D) or for krox20 alone (F-I). The insets in A-D show tailbud views of the corresponding embryos (see Fig. 3). Wild-type (WT) and krox20fh227/fh227 mutant embryos are compared in F-I. (E) Distribution of the embryos according to krox20 expression in r3 and r5 (see Fig. 3). **, P<0.0001; χ2-test. EXPRESSION / LABELING:
PHENOTYPE:
|
|
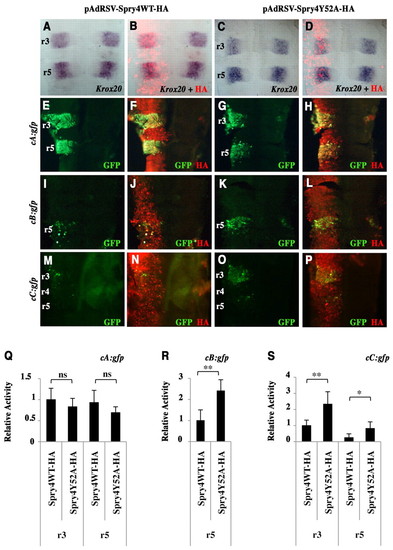
Spry4 regulates Krox20 initiator enhancers in the chick embryo. (A-D) Chick embryo neural tubes were electroporated on the left side with HA-tagged wild-type or Y52A dominant-negative zebrafish spry4-expressing vectors (pAdRSV-Spry4WT-HA and pAdRSV-Spry4Y52A-HA), and flat-mounted after in situ hybridisation with a Krox20 probe. No difference was detected between the left (experimental) and right (control) sides. The efficiency of electroporation and spry4 expression were monitored by immunolabelling against the HA tag (B,D). (E-P) Chick embryo neural tubes were co-electroporated with HA-tagged wild-type or Y52A dominant-negative spry4-expressing vectors and constructs carrying chicken Krox20 enhancer elements cA, cB or cC driving the gfp reporter, and subjected to HA (red) and GFP (green) immunostaining (merge in yellow). (Q-S) Quantitative evaluation of relative reporter activity obtained by dividing the GFP signal intensity by the HA signal intensity, both quantified using ImageJ. ns, not significant; *, P<0.0005; **, P<0.0001; Student′s t-test. Errors bars indicate s.e.m. |
|
FGF signalling is required for krox20 enhancer B activity in the zebrafish embryo. (A-D) Transgenic embryos carrying the gfp gene under the control of the chicken Krox20 A enhancer [Tg(cA:gfp)] (A,B) or the zebrafish krox20 B enhancer [Tg(zB:gfp)] (C,D) were incubated in DMSO carrier (A,C) or SU5402 (B,D) from the 1-somite stage, collected at the 8-somite stage and subjected to double in situ hybridisation for gfp (blue) and krox20 (orange); overlap is purple (A-C). Embryos were flat-mounted with anterior to the left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
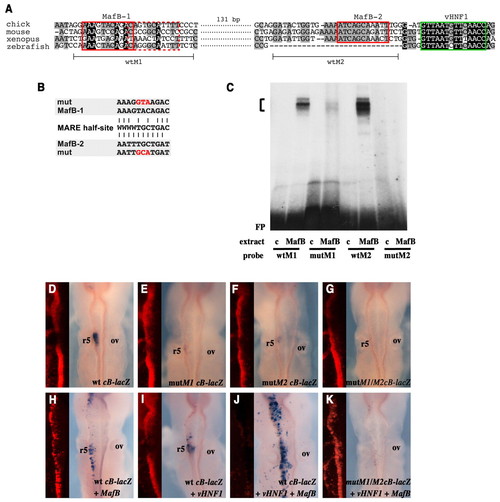
Identification of functional MafB binding sites in Krox20 enhancer B. (A) Alignment of zebrafish, Xenopus, chick and mouse Krox20 element B nucleotide sequences showing the two conserved putative MafB sites MafB-1 and MafB-2 (red boxes). A sequence of lower similarity to the MafB consensus binding site, adjacent to MafB-1 and in the reverse orientation, is also indicated (dashed red box). A vHnf1 binding site is indicated by the green box. The oligonucleotides used for gel retardation (wtM1 and wtM2) are indicated beneath. (B) Alignment of MafB-1 and MafB-2 with the consensus MafB recognition element (MARE) half-site (W=A or T). The mutations introduced into the MafB-1 and MafB-2 sites are indicated in red. (C) Gel retardation analyses were performed with the indicated bacterial protein extracts (c, control without MafB protein) and oligonucleotides carrying the chick versions of the MafB-1 and MafB-2 sites, either wild-type (wtM1, wtM2) or mutated (mutM1, mutM2). FP, free probe. The bracket indicates MafB-probe shift complexes, which are abolished or largely abolished by mutation of the MafB-2 and MafB-1 sites, respectively. (D-K) Chick embryos were analysed by X-gal staining after electroporation with constructs containing wild-type (D,H-J) or mutant (E-G,K) versions of chick element B driving a β-globin promoter-lacZ reporter. Embryos were co-electroporated with MafB alone (H), vHnf1 alone (I), or both (J,K). In all cases, a Cherry-expressing vector was co-electroporated to monitor electroporation efficiency; Cherry visualisation (red) was carried out following X-gal staining and is shown to the left of each image. Note that strong X-gal staining quenches Cherry fluorescence. ov, otic vesicle. |
|
The hindbrain patterning phenotype is not affected by a p53 morpholino. (A-D) Embryos injected with either control morpholino (Ctrlmo) (A; n=12) or Spry4mo (B; n=19), and embryos co-injected with p53 morpholino and Ctrlmo (p53mo+Ctrlmo) (C; n=12) or Spry4mo (p53mo+Spry4mo) (D; n=20) were collected at the 10-somite stage and subjected to in situ hybridisation for krox20 and her5. Embryos were flat-mounted with anterior to the left. (E) Quantitative evaluation of rhombomere areas was carried out as described in Fig. 2. *, not significant after Bonferroni corrections considering eight comparisons, P>0.006; **, P<0.006; ***, P<0.0001; t-test. Errors bars indicate s.e.m. |
|
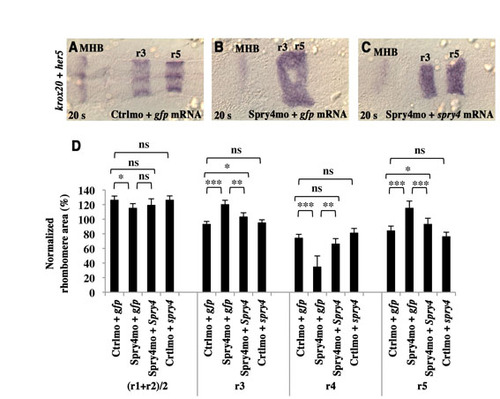
Hindbrain patterning defects resulting from Spry4mo injection are persistent and can be rescued by injection of spry4 mRNA. (A-C) Embryos were injected with either control morpholino (Ctrlmo) and gfp mRNA (A), Spry4mo and gfp mRNA (B), or Spry4mo and a morpholino-resistant form of spry4 mRNA (C), collected at the 20-somite stage and subjected to in situ hybridisation for krox20 and her5, a marker of the MHB. (D) Quantitative evaluation of rhombomere areas, carried out as described in Fig. 2. ns, not significant, P>0.05; *, not significant after Bonferroni corrections considering 12 comparisons, P>0.004; **, P<0.004; ***, P<0.0001; t-test. Errors bars indicate s.e.m. |
|
Cell proliferation in the hindbrain is not affected in Spry4 morphants. (A,B) Immunostaining against phospho-Histone H3 (phospho-HH3, green) was carried out following fluorescent in situ hybridisation using a krox20 probe (red) in 5-somite stage embryos injected with control morpholino (A) or Spry4mo (B). The images are z-projections of nine confocal sections. (C) Normalised proliferation indexes were obtained by dividing the average number of phospho-HH3-positive cells per confocal section in a given rhombomere by the area of the corresponding rhombomere. The ratio obtained in r3 with the control was arbitrarily set at 1, to which the others were then normalised. No significant difference was detected by t-test, P>0.05. Errors bars indicate s.e.m. |
|
Enhanced FGF signalling in Spry4 morphants. (A) Enhanced ERK1/2 phosphorylation in Spry4 morphants. Control and Spry4mo-injected embryos were collected at 100% epiboly and subjected to western blot analysis using antibodies against phosphoERK1/2 (pERK1/2) or total ERK1/2. Actin was used as a loading control. (B-E) Immunostaining for pERK1/2 (B,C) and in situ hybridisation for pea3 (D,E) of Ctrlmo- and Spry4mo-injected embryos at 100% epiboly. Embryos are shown as dorsal views with anterior to the top. The insets show tailbud views, allowing determination of the developmental stage by evaluation of the closure of the tailbud. tb, tailbud; hb, hindbrain; tl, telencephalon. |
|
spry4 expression in the hindbrain is dependent on FGF signalling. Embryos were incubated in either DMSO carrier (A) or SU5402 (B) from 50% to 100% epiboly and analysed at this latter stage by in situ hybridisation for spry4. Embryos are shown as dorsal views with anterior to the top. The stripe of spry4 mRNA at the level of the hindbrain is absent after SU5402 treatment, whereas tailbud expression is unaffected. tb, tailbud; hb, hindbrain; tl, telencephalon. |
|
Spry4 controls the onset of mafba expression. (A-D) Embryos injected with either control morpholino (Ctrlmo) (A,C) or Spry4mo (B,D) were collected at 100% epiboly and subjected to in situ hybridisation with mafba and ntl probes (A,B) or an fgf8 probe (C,D). The arrow in A indicates mafba expression in only a few r5/r6 cells. Embryos are shown as dorsal views with anterior to the top. The insets show tailbud views, allowing determination of the developmental stage by evaluation of the closure of the tailbud, as revealed by ntl (A,B) or fgf8 (C,D) expression. |
|
Synergy between vHnf1 and MafB for the activation of endogenous Krox20 in chick. Chick embryos were analysed by in situ hybridisation for Krox20 after electroporation with expression vectors for (A) vHnf1, (B) MafB or (C) both. Hindbrains were flat-mounted with anterior to the top. With vHnf1, limited ectopic Krox20 expression occurs specifically in r4. With MafB, limited ectopic activation of Krox20 is observed in the posterior hindbrain. With both expression vectors together, ectopic expression occurs more frequently and throughout the entire hindbrain. |