- Title
-
The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b
- Authors
- Valentin, G., Haas, P., and Gilmour, D.
- Source
- Full text @ Curr. Biol.
|
Loss of SDF1a Leads to a Stronger Phenotype than Loss of Cxcr4b (A) Neuromast deposition was detected at 4.5 days postfertilization (dpf) by alkaline phosphatase staining. Wild-type embryos show a zebrafish-specific neuromast pattern (upper panels, overview and magnified view of dashed box). In medusa mutants, the neuromast number in the posterior lateral line is reduced (arrowheads), whereas anterior neuromasts are still present (asterisk) (lower panels, overview and magnification of dashed box). (B) Schematic representation of a full-length SDF1a protein in the wild-type and a mutated SDF1a protein in medusa mutants (the colors represent the four exons of the sdf1a gene). A point mutation at nucleotide position 99 converts phenylalanine into a stop codon (red arrow). This nonsense mutation truncates SDF1a after 32 amino acids in medusa mutants. (C) The lateral-line-primordium migration was analyzed in wild-type, cxcr4b, and sdf1a mutant embryos at 42 hpf. Six equal sections from the ear to the end of the tail were defined, and an equal number of embryos (n = 37) were classified according to the leading-edge position of the primordium (arrows). (D) Quantification of results from (C) shows that SDF1a mutants have a stronger migratory defect than do cxcr4b mutants (compare red and yellow bars). |
|
The Zebrafish Homolog of Cxcr7 Rdc1 Is Essential for Primordium Migration (A) Fluorescent in situ hybridization of Cxcr7 and Cxcr4b in the wild-type CldnB::lynGFP background at 36 hr postfertilization. The middle panel shows the restricted expression pattern of Cxcr7 at the back of the primordium and in the deposited chain cells. The lower panel reveals Cxcr4b expression in the whole primordium. (B and C) Quantification of primordium migration (as in Figure 1C) in wild-type embryos, wild-type embryos injected with control morpholino (Control Cxcr7Mo, 1 mM), or two morpholinos against Cxcr7 (Cxcr7Mo1, Cxcr7Mo2, 1 mM), and cxcr4b embryos injected with Cxcr7Mo1. Wild-type and control morpholino-injected embryos migrate at a similar rate (blue and yellow bars). Primordia treated with the two morpholinos against Cxcr7 (red and purple bars) stop preferentially in part 2; only a few reach parts 3 and 4 or halt already in sector 1. Injection of Cxcr7Mo1 in cxcr4b embryos recapitulates the sdf1a mutant phenotype (green bars, compare Figure 1D). |
|
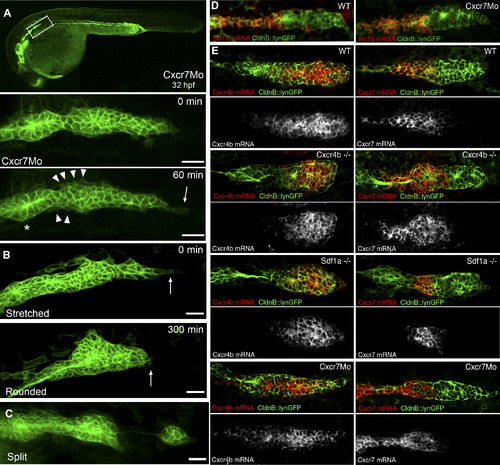
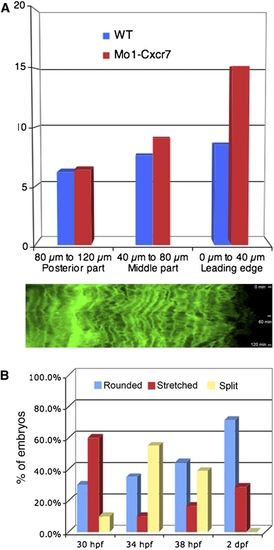
Phenotypic Analysis of Primordium Behavior after Cxcr7 Knockdown (A) Representative example of a Cxcr7 morphant primordium of the stretched class. The upper panel shows a 10× overview at 32 hpf. Below: frames taken from a 1 hr time-lapse movie (40×) that shows a stretched primordium, which does not migrate. The asterisk indicates the deposited cells, the arrowheads show cell motility at the rear of the primordium, and the arrow depicts the stretching of the leading cells. The scale bar represents 20 μm. A kymograph confirms that tip cells dynamically elongate and retract and indicates that cells within the tissue are motile but do not move (Figure S5A). (B) Time-lapse analysis of a Cxcr7 morphant embryo at 32 hpf showing the conversion from a stretched to a rounded primordium in 300 min. The arrows show the behavior of the leading cells.The scale bar represents 20 μm. (C) Example of a Cxcr7 morphant embryo at 34 hpf where primordium splits. The scale bar represents 20 μm. (D) Fluorescent in situ hybridization of krt15 in wild-type and Cxcr7 morphants at 36 hpf. Expression of krt15 is restricted to the rear of the primordium and to the deposited chain cells in both cases. (E) In situ hybridization of Cxcr4b (left panel) and Cxcr7 (right panel) in wild-type, cxcr4b mutant, sdf1a mutant, and Cxcr7 morphant embryos at 36 hpf. Although the morphology of the primordium varies between genotypes, it is clear that the relative expression domains of Cxcr4b and Cxcr7 are unaffected in the different backgrounds. |
|
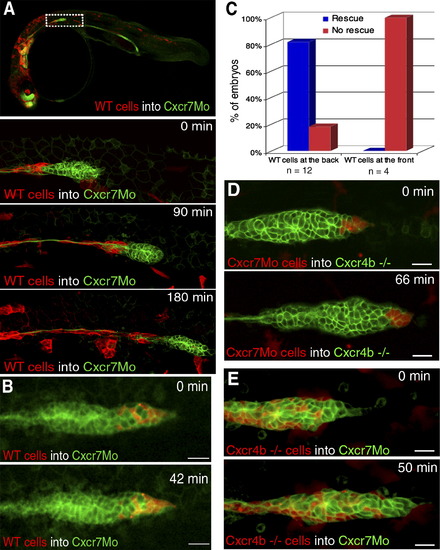
Genetic Mosaic Analysis Reveals Spatially Restricted Requirements for Cxcr7 and Cxcr4b during Lateral-Line Primordium Migration (A) A 10× overview of wild-type cells (red labeled, lyn-dtTomato mRNA) transplanted into a Cxcr7 morphant embryo at 34 hpf. Time-lapse analysis of a Cxcr7 morphant primordium displays a contiguous clone of wild-type cells in the trailing region. Migration of the primordium is efficiently rescued only up until the point when wild-type cells are deposited. The scale bar represents 20 μm. (B) Time-lapse analysis of a Cxcr7 morphant revealing that wild-type cells transplanted at the leading edge of the primordium cannot rescue the migration of a Cxcr7 morphant embryo. The scale bar represents 20 μm. (C) Embryos were screened for the localization of wild-type cells inside the morphant primordia (at the back or at the front). Migration of rescued primordia similar to that of the wild-type was observed in those cases with large clones of cells at the back but not at the front of the primordium. (D) Time-lapse analysis shows that clones of Cxcr7 morphant cells can efficiently rescue migration of cxcr4b mutant primordia. Donor embryos were kept to ensure that Cxcr7 was efficiently knocked down in transplanted cells. The scale bar represents 20 μm. (E) Rhodamine labeled cxcr4b mutant cells were transplanted into Cxcr7 morphants. Transplanted wild-type cells rescue the migration defect of Cxcr7 morphant primordia when present in the trailing regions of the primordium. The scale bar represents 20 μm. |
|
SDF1a Activity Is Affected in medusa Mutant Embryos (A) Rhodamine-labeled wild-type cells transplanted into a medusa mutant. Wild-type cells at the front of the primordium cannot rescue the migration deficiency. Tumbling of the tissue even causes wild-type cells to lose the leading position (arrowheads). The scale bar represents 20 μm. (B) medusa mutant primordia at 36 hpf respond to an ectopic clone of SDF1aoverexpressing cells (lower panel), whereas no displacement of the mutant tissue is observed in the absence of externally provided SDF1a (n = 50). The scale bar represents 100μm. |
|
Expression Pattern of Cxcr7 Alkaline Phosphatase in situ hybridization of Cxcr7 in a wild-type embryo at 36 hr postfertilization. A magnified view of Cxcr7 expression in the lateral-line primordium is shown in the upper-right panel. |
|
Expression Pattern of the Posterior Marker krt15 in Mutant and Morphant Backgrounds Fluorescent in situ hybridization of krt15 at 36 hpf in wild-type, sdf1a mutant, and cxcr4b mutant embryos. Lower panels show a defined expression pattern of krt15 at the rear of the primordium and in the deposited chain cells. Trailing to primordium in SDF1a and cxcr4b mutants is the posterior lateral-line ganglion, where the gene is not expressed. |
|
Cxcr7 Morpholino Specifically and Efficiently Blocks Translation of Its Target mRNA One-cell-stage embryos were injected with 60 ng/μl lyn dtTomato mRNA and either 60 ng/μl morpholino-resistant (A and B) or 60 ng/μl morpholino- sensitive (C and D) Cxcr7-GFP mRNA. Coinjection of 1 mM Cxcr7Mo did not affect expression of Cxcr7-GFP in the MO-resistant mRNA (B) but did efficiently block translation of the MO-senstitve Cxcr7-GFP sequence (D). Levels of lyn-dtTomato expression were not affected in either case. |
|
Phenotypic Analysis of Cxcr7 Morphants PHENOTYPE:
|