- Title
-
Pre-gastrula expression of zebrafish extraembryonic genes
- Authors
- Hong, S.K., Levin, C.S., Brown, J.L., Wan, H., Sherman, B.T., Huang, D.W., Lempicki, R.A., and Feldman, B.
- Source
- Full text @ BMC Dev. Biol.
|
Expression of genes in the E-YSL. Whole mount in situ hybridizations showing E-YSL expression of mRNAs encoding the transcription factors gata3 [48], znf503 [49-51], hnf1ba [52], gata6 [30] and cebpa [30,53], one cytoskeletal protein that is potentially associated with the inner plasma membrane (camsap1l1 [54]), two solute carriers (slc26a1 [30] and slc40a1 [30]), one G-protein coupled receptor (gpr137bb [55]) and one integral membrane protein (otop1 [31,56,57]). Lateral views are presented throughout, with the dorsal side (where apparent) to the right for shield and 80% epiboly stages. Developmental stages and genes probed are indicated. Arrows point to E-YSL expression and arrowheads point to I-YSL expression. Asterisks on the bottom right corner indicate genes for which no in situ expression data has previously been reported. EXPRESSION / LABELING:
|
|
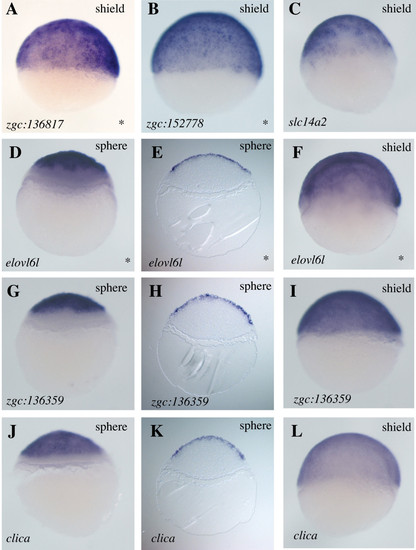
Expression of genes in the EVL. Whole mount in situ hybridizations for mRNAs expressed in the EVL encoding one RhoGEF (zgc:136817 [58]), one protein with a LIM zinc binding domain (zgc:152778 [58]), one solute carrier (slc14a2 [59]), one protein related to a family of long chain fatty acid-elongating enzymes (elovl6l [58]), one protein with a DEP domain (zgc:136359 [31]), indicating it might be involved in G-protein coupled receptor recognition, and one intracellular chloride channel protein (clica [31]). Expression of three of these genes was excluded from the margin at the sphere stage of development (D, G and J), also seen in histological sections (E, H and K). By shield stage, expression is apparent throughout the EVL (F, I and L). Lateral views are shown throughout and developmental stages and genes probed are indicated. Asterisks on the bottom right corner indicate genes for which no in situ expression data has previously been reported. |
|
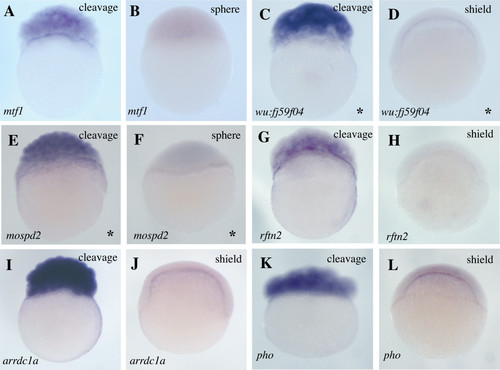
Expression of transient maternal genes. Whole mount in situ hybridizations showing what we presume to be maternal expression for mRNAs encoding one transcription factor (mtf1 [60]), two proteins with no recognizable motifs (wu:fj59f04 and pho [61]), one protein with similarity to the human MOTILE SPERM DOMAIN CONTAINING 2 gene (mospd2), one protein belonging to a lipid raft-linking family (rftn2 [31]) and one protein with an arrestin domain (arrdc1a [30]). Lateral views are shown and developmental stages and genes probed are indicated. Asterisks on the bottom right corner indicate genes for which no in situ expression data has previously been reported. |
|
Nodal-independent regulation of genes in the YSL. Whole mount in situ hybridizations on WT and MZoep embryos at the early-gastrula (6 hpf) stage. Lateral magnifications of the margin and YSL are shown throughout, except for lateral whole-embryo views in M′, N′, O′ and P′. No significant difference was seen in the YSL expression levels of mxtx1 in WT or MZoep embryos, as was previously reported (A-B). Similarly, no significant difference was seen between WT and MZoep embryos for the YSL expression of hnf4a (C-D), gpr137bb (E-F), or camsap1l1 (G-H). The expression of lhx1b (I-J) and sp5 (K-L) was substantially reduced in MZoep mutants. Similarly, concentrated expression of zgc:110712 near the margin was absent in MZoep embryos (M, M′, N, N′). The gata6 gene is expressed both in the YSL and margin (O and O′) and the margin expression domain is eliminated in MZoep mutants, whereas the YSL expression domain persists (P and P′). EXPRESSION / LABELING:
|
|
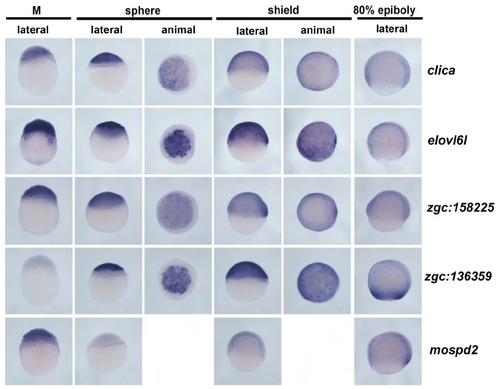
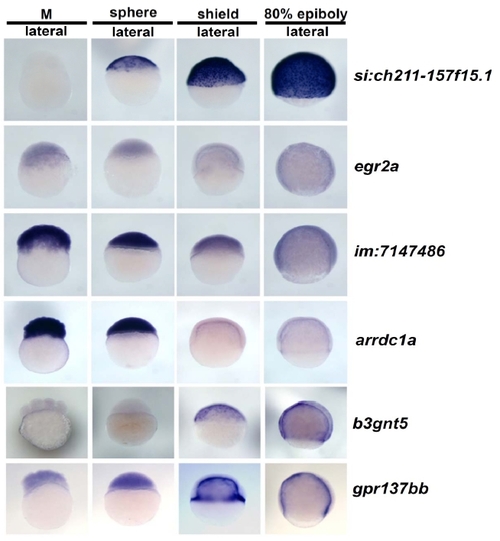
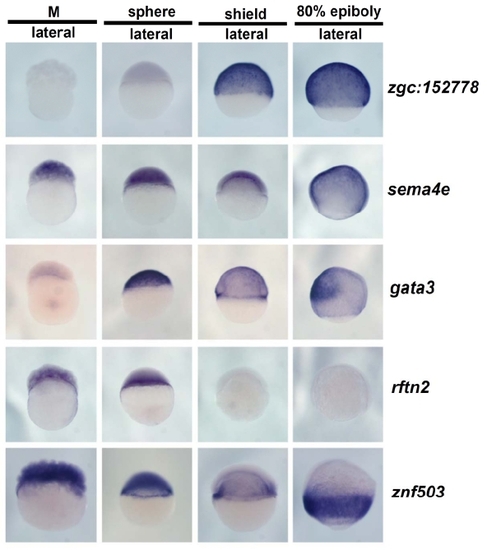
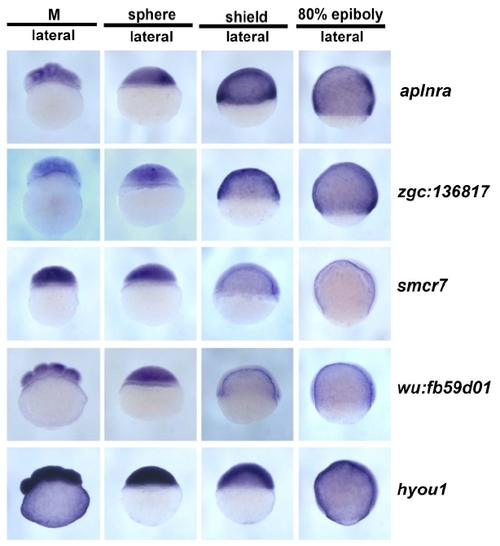
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |
|
Whole-mount in situ hybridization images for 56 genes. Two hundred and forty images representing four stages with some multiple views for each of fifty-six genes are presented on 11 pages (A through K). The location of data for particular genes is indicated in Additional File 1. No data is shown for ten genes for which no expression was detected and two genes that showed ubiquitous expression. |