- Title
-
Chemokine signaling guides regional patterning of the first embryonic artery
- Authors
- Siekmann, A.F., Standley, C., Fogarty, K.E., Wolfe, S.A., and Lawson, N.D.
- Source
- Full text @ Genes & Dev.
|
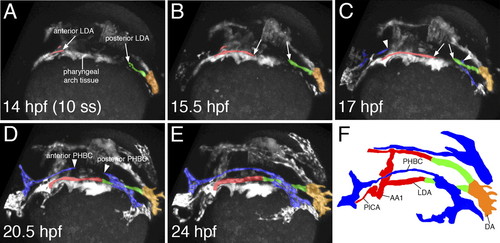
Two-photon time lapse of LDA formation in live zebrafish embryos. (A–F) Dorsolateral view of LDA formation. Anterior LDA is pseudocolored in red, posterior LDA is shown in green, axial DA is indicated by orange, and the PHBC is shown in blue. Arrows mark migrating cells of the LDA, which forms in a bidirectional manner. Arrowheads indicate similarly migrating PHBC cells. (F) Labeled camera lucida image indicating position of vessels in E. (DA) Dorsal aorta; (LDA) lateral dorsal aorta; (PHBC) primordial hindbrain channel; (PICA) primitive internal carotid artery; (AA1) aortic arch 1. EXPRESSION / LABELING:
|
|
LDA progenitors are genetically distinct. (A–H) In situ hybridization with indicated markers. (A–D,G) Dorsal view, anterior is up. (E,F,H) Lateral view, anterior to the left, dorsal is up. (A–D) Expression of cxcr4a (A,C) and kdrl (B,D) at the 18-somite stage and the 22-somite stage, respectively. Anterior LDA (black arrows) and posterior LDA (white arrowheads) are indicated. Black line marks the anterior end of the notochord. Asterisks in C mark expression of cxcr4a in pharyngeal arch tissue. (E,F) Confocal images of 22-somite-stage embryos following double-fluorescent in situ hybridization. Endothelial cells are labeled by expression of fli1a in red and cxcr4a in green. (G,H) Expression of vsg1 at the 22-somite stage. (G) Dorsal view. (H) Lateral view. (G) Black arrows mark posterior LDA, which does not express vsg1. Bracket marks anterior end of DA. Blue arrowheads mark venous cells. (I) Schematic drawing of DA domains delineated by different migratory behaviors and gene expression profiles at the 18-somite stage and the 22-somite stage. EXPRESSION / LABELING:
|
|
LDA formation defects in cxcr4a-deficient embryos. (A) Schematic depicting ZFN monomers designed against cxcr4a exon 2 target site. Nucleotides in pink represent the recognition elements. The DdeI site within the spacer region is underlined. Recognition helices for each finger are indicated. (B) PCR genotyping analysis of embryos derived from putative founder fish. Failure to digest with DdeI is indicative of a possible target site deletion. An asterisk marks genotypes that bear mutagenic lesions. (C,F) Transmitted light images of wild-type (C) and cxcr4a mutant embryos (F) at the 32-hpf stage. Lateral views, dorsal is up, anterior is to the left. (D,E,G,H) Confocal images showing anterior regions (D,G) or posterior regions (E,H) of wild-type (D,E) or cxcr4a mutant (G,H) embryos. (LDA) Lateral dorsal aorta; (PHBC) primordial hindbrain channel; (aa1) aortic arch1. Bracket in G indicates a gap within LDA. (DLAV) Dorsal longitudinal anastomotic vessel; (DA) dorsal aorta; (PCV) posterior cardinal vein. The arrow in E indicates the intersegmental blood vessel sprout. (I–M) Stills of two-photon time-lapse (see Supplemental Movie 5) showing LDA formation in cxcr4aum20 mutant embryos. Lateral views, anterior is to the left. Time points are indicated. Red labels arterial LDA cells, green indicates posterior LDA cells, and blue labels venous endothelial cells. Arrows mark anterior and posterior LDA, while arrowheads mark the forming PHBC. The green arrow in M marks ectopically located endothelial cells. (N) Camera lucida drawing of embryo in M. EXPRESSION / LABELING:
|
|
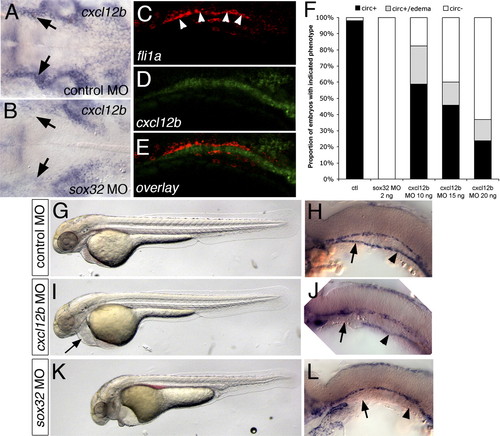
Endoderm-derived cxcl12b is required for LDA formation. (A,B) In situ hybridization at 24 hpf showing cxcl12b expression in tissue underlying the LDA (arrows). (B) Loss of endoderm in sox32 morpholino-injected embryos leads to severe reduction in cxcl12b expression. Dorsal view, anterior is to the left. (C–E) Confocal micrographs of 22-hpf embryo subjected to double-fluorescent whole-mount in situ hybridization, lateral view, anterior is to the left, dorsal is up. (C) fli1a expression, marking the LDA. (D) cxcl12b expression. (E) Overlay; fli1a is red, cxcl12b is green. (F) Influence of loss of endoderm or cxcl12b on circulation. (ctl) Control morpholino-injected; (circ+) embryos with circulation; (circ) embryos without circulation. (G–L) Vascular defects in embryos lacking endoderm or cxcl12b. (G,I,K) Bright-field views at 32 hpf, anterior is to the left, dorsal is up. (H,J,L) In situ hybridization for kdrl at 24 hpf. Black arrows indicate anterior LDA, arrowheads indicate posterior LDA. (G,H) Embryos injected with control morpholino. (I,L) Embryos injected with 15 ng of cxcl12b MO. (K,L) Embryos injected with 2 ng of sox32 morpholino. Arrows mark the anterior LDA, arrowheads mark the posterior LDA. EXPRESSION / LABELING:
|
|
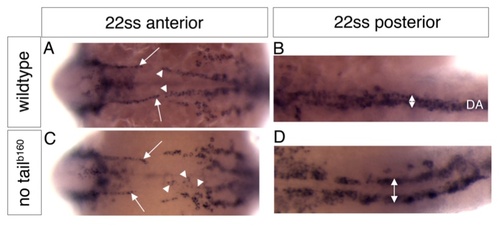
Absence of the notochord specifically affects posterior dorsal aorta development. Wildtype (A, B) or no tail b160 mutant embryos (C, D) at the 22 somite stage (ss) in situ hybridized with a probe for kdrl marking all endothelial cells. Anterior is to the left, dorsal up. A, Anterior region of a wildtype embryo showing the forming paired anterior lateral dorsal aortae (arrows), or the posterior lateral dorsal aortae (arrowheads). A, Posterior region of the same embryo as in A, showing the single posterior dorsal aorta (double arrow, DA). C, In no tail b160 mutant embryos, formation of the anterior lateral dorsal aortae occurs normally (arrows), while the posterior dorsal aortae (arrowheads indicate individual endothelial cells close to the midline) do not form properly. D, Posterior region of the same embryo as in C, showing failure of angioblasts to migrate towards the midline (double arrow, compare to double arrow in B). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Still images from 2-photon time lapse movies from wild type Tg(kdrl:egfp) la116 embryos showing endothelial cell movements contributing to lateral dorsal aorta and dorsal aorta formation. A-E, Dorso-lateral view of posterior lateral dorsal aorta formation. Time points are indicated. Endothelial cells that contribute to the lateral dorsal aortae are labeled in green; dorsal aorta is labeled in orange. Note the formation of a Y-shaped structure, consisting of the paired lateral dorsal aortae anteriorly and the single dorsal aorta posteriorly. C, sprouting intersegmental blood vessels are indicated by arrowheads. F, labeled camera lucida image indicating position of vessels in e. ISV – intersegmental blood vessel. EXPRESSION / LABELING:
|
|
Expression pattern of cxcr4a during zebrafish embryonic development. All embryos are shown with their anterior to the left, dorsal side up. A, C, E, G, side views, B, D, F, H, top views. A, B, 12 somite stage (ss;15 hours post fertilization), expression of cxcr4a can be detected in discrete cells in the anterior lateral mesoderm (arrows). C, D, 18 ss, anterior lateral dorsal aorta cells express cxcr4a (arrowheads). E, F, 22 ss, expression of cxcr4a continues in anterior lateral dorsal aorta cells (arrowheads). Strong expression of cxcr4a can also be detected in pharyngeal arch tissue (F, black arrows). G, H, 24 hpf, expression of cxcr4a can be detected in the lateral dorsal aorta (arrowhead), the head vasculature, for instance the carotid arteries (ca), and the dorsal aorta in the trunk (arrow). EXPRESSION / LABELING:
|
|
Phenotypic comparison of wildtype and cxcr4a um20 mutant embryos at 24 hpf. A-C, wildtype, E-F, cxcr4a um20 mutant. Lateral views, anterior to the left. A, D, overview of general morphology. Arrow in B indicates forming edema in cxcr4a um20 mutant embryos. B, E, close-up of axial vasculature. N-notochord, A-dorsal aorta, V-posterior cardinal vein. Black arrow indicates pooled blood cells in the posterior cardinal vein in cxcr4a um20 mutant embryos due to lack of blood flow. C, F, somite boundaries (white arrowheads) form normally in cxcr4a um20 mutant embryos. PHENOTYPE:
|
|
Endoderm patterning defects in cxcr4a mutant embryos. A-C, sox32 in situ hybridization of 90% epiboly embryos. Dorsal is to the right, animal pole up. Note anterior displacement of endodermal cells in cxcr4a um20 mutant embryos (B, C, arrows) when compared to wild type embryo (A). D-K, insulin in situ hybridization at 55 hours post fertilization in 4 representative wild type embryos (D-G), compared to cxcr4a um20 mutant embryos (H-K). Note displaced insulin expressing cells (arrowheads) in J, K. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Stills of 2-photon movies showing defects in lateral dorsal aorta formation. Timepoints are indicated. Aorta is pseudocolored in red, vein is pseudocolored in blue. Arrows mark the positions of the branches of the anterior lateral dorsal aortae (red) and the posterior lateral dorsal aortae (green), respectively. Dorsal aorta cells are pseudocolored in orange. Arrowheads mark the positions of the primordial hindbrain channel branches. C, F, I, L, camera lucida drawings. LDA-lateral dorsal aorta, PHBC-primordial hindbrain channel, DA- dorsal aorta, AA1-aortic arch 1. A-C, wildtype embryo, D-F, embryo injected with cxcr4a morpholino. G-H, sox32 morpholino (MO) injected embryo lacking endoderm. J-K, embryo injected with cxcl12b morpholino. Note the lack of dorsal aorta closure in embryos lacking cxcr4a, cxcl12b or the endoderm (sox32 morpholino injected embryos), while development of the primordial hindbrain channel proceeds normally. EXPRESSION / LABELING:
PHENOTYPE:
|
|
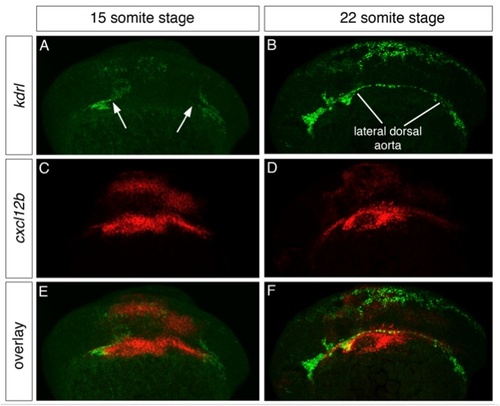
The chemokine cxcl12b is expressed prior to the formation of the lateral dorsal aorta. Double fluorescent in situ hybridization with cxcl12b (red) and kdrl (green). Dorso-lateral views, all embryos are oriented anterior to the left, dorsal up. A, C, E 15 somite stage. B, D, F 22 somite stage. A, kdrl is expressed in endothelial cells that begin to form the lateral dorsal aorta (arrows). B, cxcl12b is expressed in the region in which the lateral dorsal aorta will form. E, overlay of green and red channels. B, kdrl expression showing the fully formed lateral dorsal aorta. D, Expression of cxcl12b in the region of the formed lateral dorsal aorta. F, overlay of green and red channels. EXPRESSION / LABELING:
|