- Title
-
Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish
- Authors
- McMahon, C., Gestri, G., Wilson, S.W., and Link, B.A.
- Source
- Full text @ Dev. Biol.
|
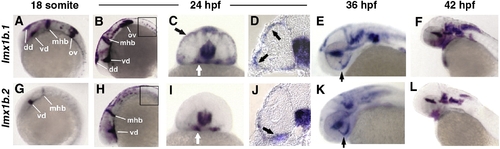
Zebrafish lmx1b.1 and lmx1b.2 are expressed in the ocular and non-ocular tissue. In situ hybridization of lmx1b.1 (A–F) and lmx1b.2 (G–L) from 18 s to 42 hpf. Anterior is to the left in lateral views. Expression of both lmx1b genes was strong in the midbrain–hindbrain boundary and ventral diencephalon at 18 s (A, G) and 24 hpf (B, H). Only lmx1b.1 was detected in the otic vesicle (A versus G) and spinal cord neurons (B versus H, boxed). For both genes, periocular expression in cells associated with the optic stalk (white arrows C, I) and the globe of the eye (black arrows C, I), was observed in whole mount and histological sections (arrows D, J). Expression of lmx1b.1 and lmx1b.2 was also detected in the ventral fissure (arrows E, K) and hindbrain neurons (E, F and K, L). See text for additional details. mhb, midbrain–hindbrain boundary; vd, ventral diencephalon; dd, dorsal diencephalon; ov, otic vesicle; hb, hindbrain. EXPRESSION / LABELING:
|
|
Lmx1b knock-down disrupts ocular morphogenesis to variable degrees. Lateral view of 24 hpf heads (A, D) and higher magnification of 36 hpf PTU-treated eyes (E, H) with anterior to the left. (A–P) Control embryos and embryos with different degree of severity of the Lmx1b morpholino phenotypes as indicated on the top of each column. (H) Dots highlight the shape of the ventral retina. (E–H) Bars indicate the choroid fissure, which fails to close in the MO-injected embryos. Transcripts for foxc1a (I–L) and eya2 (M–P) in control MO and differently affected lmx1bdMO morphant embryos as revealed by whole mount in situ hybridization at 36 hpf. Reduction of periocular cells was observed in anterior-ventral regions of moderate lmx1bdMO embryos (asterisk in panel O) and completely absent in severe morphants (L, P). Toluidine blue-stained transverse (Q, R) and sagittal (S, T) sections from control (Q, S) and moderate lmx1bdMO phenotypes (R, T) at 24 hpf and 62 hpf, respectively. |
|
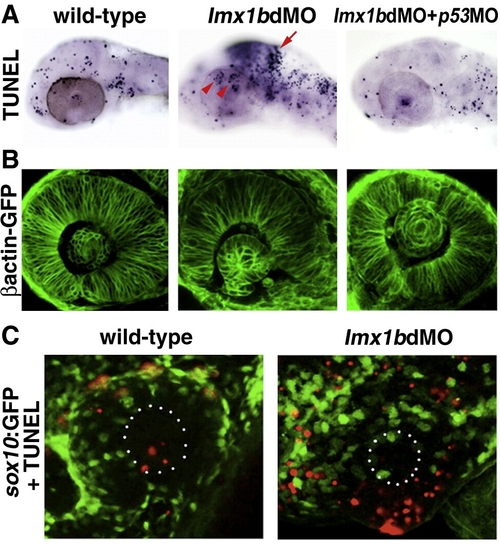
Lmx1b loss of function induces apoptosis of Lmx1b expressing cells. (A) TUNEL staining of apoptotic cells (dark blue, top row) in lmx1bdMO morphants and lmx1bdMO morphants co-injected with p53MO and analyzed at 36 hpf. Red arrow indicates increased apoptosis in the midbrain–hindbrain boundary. Red arrowheads show increased periocular cell death in lmx1bdMO eyes. (B) Eye morphology in embryos corresponding to the above panel as visualized in the betaactinGFP transgenic line. (C) TUNEL staining (red) in sox10:GFP-positive neural crest cells (green) in wild-type and lmx1bdMO morphant eyes. White dots in B outline the lens. |
|
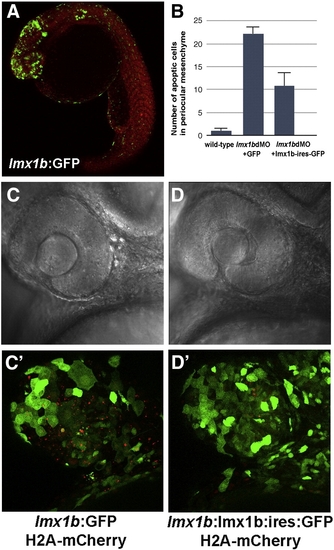
Apoptosis in lmx1bdMO morphants is rescued by expression of lmx1b.1 cDNA. (A) Expression of GFP driven by the - 5 kb lmx1b.1 promoter. (B) Quantitation of the number of pyknotic cells present in ocular regions from wild-type, lmx1bdMO morphants expressing GFP, and lmx1bdMO embryos expressing lmx1b.1:ires:GFP. For each condition, n = 10 independent eyes were scored. Error bars represent Standard Error of the Mean. (C) Bright field image of lmx1bdMO morphants expressing GFP or (D) lmx1bdMO morphants expressing lmx1b.1:ires:GFP. (C′) Fluorescent image of panel C showing lmx1b:GFP-positive periocular cells (green) and pyknotic cells red). (D′) Fluorescent image of panel D showing lmx1b:lmx1b.1:ires:GFP-positive periocular cells (green) and pyknotic periocular cells (red). |
|
Apoptosis and proliferation in lmx1b.1:GFP-positive cells following loss of Lmx1b function. (A) Immunoreactivity for activated Caspase-3 (red) and fluorescence for lmx1b.1:GFPmw11 (green) in control MO or (B) lmx1bdMO ocular cryosections at 36 hpf. Arrows indicate cells positive for both activated Caspase-3 and lmx1b.1:GFP, while arrows indicate GFP-negative apoptotic cells. (C) Immunoreactivity for phosphoHistone (red) and fluorescence for lmx1b.1:GFPmw11 (green) in control MO or (D) lmx1bdMO ocular cryosections at 36 hpf. Arrows indicate cells positive for both activated phosphoHistone-3 and lmx1b.1:GFP, while arrows indicate GFP-negative mitotic periocular cells. Dotted lines indicate position of the lens. Images shown are from representative sections (n = 24 total embryos for each condition from 2 experiments). |
|
Lmx1b functions cell-autonomously in development of periocular mesenchymal cells. (A) Bright field and fluorescent images of control MO and lmx1bdMO eyes showing foxD3:GFP-positive periocular mesenchymal cells. (B) Ocular region (outlined by white dots) of 30 hpf mosaic embryos showing wild-type or lmx1bdMO morphant donors. All donor cell nuclei are positive for H2A:mCherry (red), while a subset of periocular cells are positive for foxD3:GFP (green). (C) Average number ± Standard Error of the Mean for total H2A:mCherry and foxD3:GFP-positive donor periocular cells from mosaic embryos. Percent of total periocular cells that were foxD3-positive is shown in the right-most column. For each condition, donor cells from n = 12 independent eyes were scored. EXPRESSION / LABELING:
|
|
Migration of lmx1b.1:GFP-positive cells is altered following knock-down of Lmx1b. Bright field and fluorescence images of (panels A–C) control MO and (panels D–F) lmx1bdMO embryos from the Tg(- 5 kb lmx1b.1:GFP)mw11 line. For each image developmental time is indicated at the top, anterior is to the left, and dorsal is up. Arrows indicate the location of the optic stalk region. (D) Note the significant reduction of GFP fluorescence in Lmx1b-deficient cells associated with the optic stalk. Each fluorescent image is a compressed z-stack, which includes the entire globe of the eye and optic stalk region. Images shown are from representative embryos (n = 12 total embryos tracked over time from 2 experiments). EXPRESSION / LABELING:
PHENOTYPE:
|
|
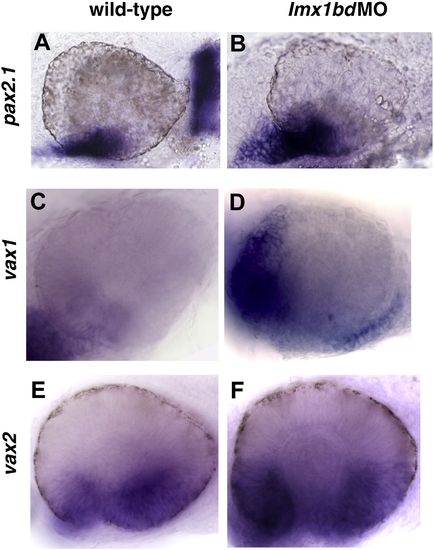
Lmx1b loss of function promotes the expansion of ventral ocular markers. Transcripts for pax2.1 (A, B), vax1 (C, D), vax2 (E, F) detected by whole mount in situ hybridization in uninjected control and lmx1bdMO embryos at 24 hpf. EXPRESSION / LABELING:
|
|
Fgf signaling is activated in lmx1b morphant ocular regions. Whole mount in situ hybridization for fgf3 (A–D) and fgf8 (E–H) in control and lmx1bdMO embryos showing head (A, B, E, F) and higher magnification of the eyes (C, D, G, H). Note the expansion of fgf3 and fgf8 in optic stalk and ventral retinal tissues of lmx1bdMO embryos (solid arrows in panels B, F). In contrast, midbrain–hindbrain expression was decreased in lmx1bdMO morphant embryos (open arrows in panels B, F). EXPRESSION / LABELING:
|
|
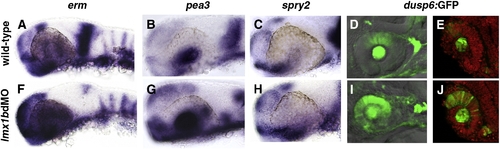
Expression of Fgf-target genes is altered in lmx1b morphants. Whole mount in situ hybridization in 24 hpf embryos for erm, pea3, and spry2 (A–C, F–H) and fluorescence in Tg(dusp6:dEGFP) (D, E, I, J). For each Fgf-target gene, note the expansion in periocular and ocular regions and reduction in the MHB. For dusp6:GFP, sections confirmed up-regulation of GFP in the dorsal neural retina, lens, and optic stalk regions (E, J). Sections were counter-stained with propidium iodide to highlight nuclei (red). EXPRESSION / LABELING:
|
|
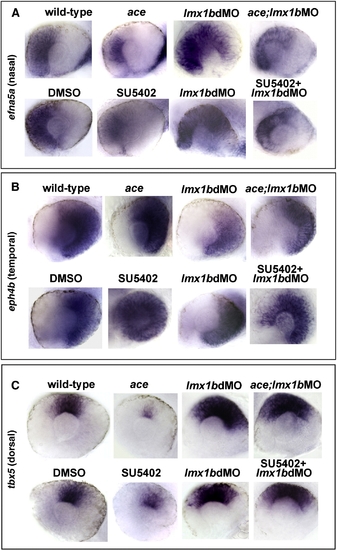
Lmx1b is required for patterning the retina in a manner partially independent from Fgf signaling. Expression of nasal, temporal, and dorsal ocular markers with reduced Fgf signaling and Lmx1b function. Lateral view of 24 hpf eyes from whole mount in situ hybridization of (panel A) efna5a (nasal), (panel B) eph4b (temporal), and (panel C) tbx5 (dorsal). Note the reduction of efna5a and tbx5 markers in fgf8a/ace mutants or SU5402-treated embryos and expansion of these markers in lmx1b morphants. Partially rescued and intermediate phenotypes were observed when Fgf signaling was reduced in lmx1b morphants. With eph4b, expansion was noted in fgf8a/ace mutants or SU5402-treated embryos, while the opposite was found in lmx1b morphants. Again, intermediate phenotypes were observed when Fgf signaling was reduced in lmx1b morphants. |
|
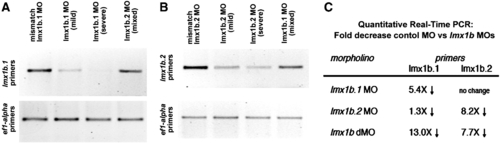
Analysis of transcript levels of lmx1b.1 and lmx1b.2 following morpholino knock-down. Ethidium bromide stained gel of RT-PCR analysis of either (A) lmx1b.1 or (B) lmx1b.2. For each, the cDNA source is shown at the top and amplification from ef1-alpha primers is shown as an input control. With either lmx1b.1 or lmx1b.2 morpholinos, we did not detect alternate splice bands, but did find depletion of the normally-spliced transcript. For lmx1b primers 35 cycles were carried out, while reactions with ef1-alpha were amplified for 20 cycles. Mild and Severe refers to the degree of the phenotype as defined in the Materials and methods. Mixed indicates equal numbers of mild and severe. (C) Fold change in transcript levels from 18 somite stage embryos as assessed by quantitative real-time RT-PCR. |
|
Vascular patterning is disrupted in the eyes of lmx1b morphants. Fluorescence from GFP expressed under the fli1 regulatory sequence (Tg(fli1a:EGFP)y5 ; Lawson and Weinstein, 2002). Lateral views of the eye show vascular endothelial cells in wild-type embryos (A) and two examples from lmx1b morphants (B, C). |
|
lmx1b.1:GFP fluorescence is lost in the optic stalk region, but maintained in the mid-ventral diencephalon of lmx1bdMO embryos. (A, D) Bright field images of control or lmx1bdMO embryos for the Tg(- 5 kb lmx1b.1:GFP)mw13 line. (B, E) GFP-fluorescent images of the same embryos. (C, F) Combined bright field and fluorescent images. Arrow indicates the mid-ventral diencephalon, and arrowhead indicates optic stalk region. Asterisk indicates lack of GFP expression in the optic stalk region of lmx1bdMO embryos. |
|
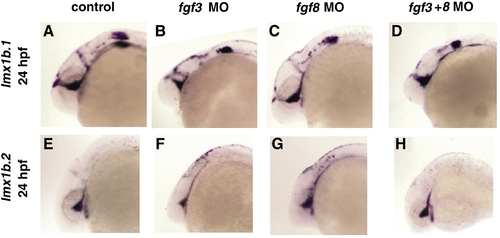
Expression of lmx1b genes in fgf morphants. In situ hybridization of lmx1b.1 (A–D) or lmx1b.2 (E–H) expression in uninjected control, fgf3 morphants, fgf8 morphants, or fgf3+fgf8 morphants at 24 hpf. No significant changes in gene expression were noted in ocular regions, although differences were found in hindbrain structures where Fgf3 and Fgf8 are required for morphogenesis. |

Unillustrated author statements EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 332(2), McMahon, C., Gestri, G., Wilson, S.W., and Link, B.A., Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish, 287-298, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.