- Title
-
Identification of direct T-box target genes in the developing zebrafish mesoderm
- Authors
- Garnett, A.T., Han, T.M., Gilchrist, M.J., Smith, J.C., Eisen, M.B., Wardle, F.C., and Amacher, S.L.
- Source
- Full text @ Development
|
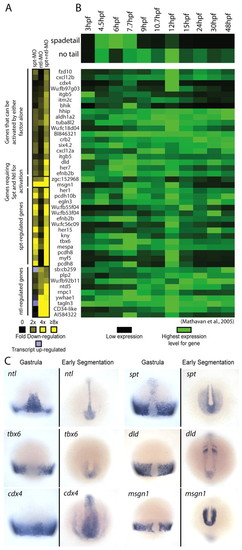
Microarray analysis identifies candidate spt and ntl target genes. (A) Gene expression levels in MO-injected embryos relative to wild-type embryos at 75% epiboly. Downregulation is shown in shades of yellow (2 to >8-fold) and upregulation (2 to 3-fold) is shown in light purple. Only genes with a two-fold or greater decrease in expression level are shown. (B) spadetail (spt) and no tail (ntl) gene expression profiles between 3 and 48 hpf are shown (top) with the expression profiles of putative targets below. These gene expression profiles were retrieved from another dataset (Mathavan et al., 2005). The expression level for each gene was normalized to its highest expression level over the time course. (C) mRNA expression of spt, ntl and four targets at midgastrula (75% epiboly, 8 hpf) and early segmentation (11-13 hpf). These patterns have been previously reported: ntl (Schulte-Merker et al., 1992), spt (Griffin et al., 1998), tbx6 (Hug et al., 1997), dld (Haddon et al., 1998; Hans and Campos-Ortega, 2002), cdx4 (Joly et al., 1992) and msgn1 (Yoo et al., 2003). EXPRESSION / LABELING:
|
|
Spt and Ntl bind similar sites in vitro. (A,B) Representative gel shifts using recombinant Spt-GST (A) and Ntl-GST (B). (C,D) WebLogo (Crooks et al., 2004) binding site models based on the sequences of >70 bound oligos from the in vitro selection assays are shown for Spt-GST (C) and Ntl-GST (D) with the count matrices for the sequenced binding sites below. Regions of binding sites contained in the non-random primer region of the oligonucleotide were not counted, therefore some columns have fewer total counts. |
|
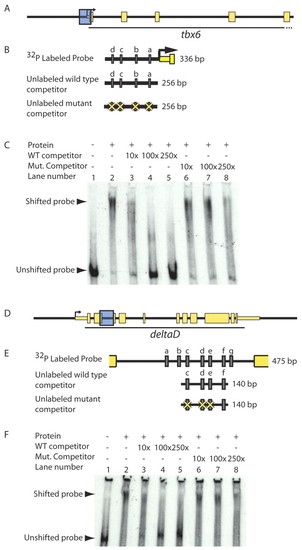
T-box proteins bind putative regulatory sequences in vitro in a T-box binding site-dependent manner. (A-F) To test whether Spt and Ntl bind putative regulatory sequences in vitro, we performed gel shifts using equal volumes of recombinant Spt-GST and Ntl-GST, and radiolabeled DNA corresponding to putative tbx6 and dld genomic regulatory sequences (shown as blue boxes in A and D and magnified in B and E to indicate Spt/Ntl binding motifs a-d and a-g, respectively, as grey rectangles). Spt and Ntl bind a 336 bp region containing four T-box sites just upstream of tbx6 (C; lane 1 versus lane 2). Addition of 10-250x excess unlabeled competitor competes well for binding and virtually eliminates DNA-protein complex formation (C; lanes 3-5); however, excess unlabeled competitor containing mutated T-box sites competes poorly (C; lanes 6-8). Similarly, a mixture of equal volumes of Spt-GST and Ntl-GST binds a 475 bp fragment from the dld second intron containing seven putative T-box sites (F, lane 1 versus 2). Unlabeled competitor containing four of the T-box sites competes for binding (F; lanes 3-5), but fails to compete when three of the T-box sites are mutated (F, lanes 6-8). |
|
The tbx6 regulatory element drives mesodermal gene expression and requires Spt and Ntl for activity. The 1.7 kb region upstream of tbx6 is shown with gray rectangles representing predicted Spt- and/or Ntl-binding site motifs (A). This region drives reporter expression at the margin at mid-gastrulation (B), but not if embryos are co-injected with spt and ntl MOs (C) (mCherry, red; GFP, green). (D-G) Endogenous tbx6 expression is decreased in spt- (F) or ntl-depleted (E) embryos when compared with expression in wild-type embryos (D). tbx6 expression is undetectable when both ntl and spt are depleted (G). |
|
T-box sites are required for activity of the tbx6 mesodermal regulatory region. (A) The 1.7kb region upstream of tbx6 is shown with gray rectangles representing predicted Spt- and/or Ntl-binding site motifs. (B) A detailed view of 300 bp upstream of the tbx6 transcription start site [gray rectangles indicate putative T-box sites (a-d) and diamonds indicate Tcf/Lef-binding sites important for regulatory element function (Szeto and Kimelman, 2004)]. (C,D) mCherry expression driven by the full-length construct is robust at mid-gastrulation (8 hpf) (C) and mid-segmentation (14 somites) (D). (E-P) During gastrulation, mCherry expression driven by constructs in which one T-box site has been mutated (E,H,K,N) is reduced (F,I,L,O), but is normal during segmentation (G,J,M,P). (Q-S) When all four sites are mutated (Q) expression is abolished at both stages (R,S). The percentages of embryos with robust, faint or no mCherry expression are given in Table 1. |
|
The second intron of deltaD contains a conserved cluster of T-box sites required for T-box-dependent mesodermal expression. (A) The second intron from fish dld orthologs (there is an unsequenced gap in the Medaka intron). Grey rectangles represent T-box binding motifs, diamonds indicate putative Tcf/Lef sites and grey ovals represent putative Suppressor of Hairless-binding sites. (B) An alignment of the second intron of fish dld orthologs reveals two particularly well-conserved Spt/Ntl-binding motifs (sites c and d). (C-F) A construct containing 1.2 kb upstream and 1.3 kb downstream sequences relative to the dld transcription start site, including the known neural regulatory element (HII) (Hans and Campos-Ortega, 2002) and the T-box site cluster (C,D) drives robust mCherry expression in the non-axial margin during gastrulation (E) and in the tail bud and somites during segmentation (F). (G-I) When two well-conserved T-box sites (c,d) and a T-box consensus sequence (e) are mutated (G), mCherry expression is greatly reduced (H,I). The percentages of embryos with robust, faint or no mCherry expression is given in Table 1. |
|
The mesodermal deltaD regulatory element requires spt and ntl function. (A-I) Stable transgenics carrying the dld reporter construct (A) express mCherry at the blastoderm margin during mid-gastrulation (G), and in the tail bud and hindbrain (C,D,H,I). Transgene expression begins by 30% epiboly (E,F; animal pole views). (J-M) Stable dld transgenics were injected with MOs targeting ntl, spt or both transcripts, and analyzed by whole-mount in situ hybridization for mCherry. Most uninjected control (J) and Ntl-depleted embryos (K) expressed mCherry [76% (38/50) and 72% (26/36), respectively]. Only 67% (22/33) of Spt-depleted embryos expressed mCherry, and only 9% (3/33) did so at a level comparable with that of uninjected controls (L). No spt+ntl MO-injected embryos (0/54) expressed mCherry (M). (N-Q) In situ hybridization revealed that endogenous dld responds to Spt and Ntl depletion in a similar manner to the transgene. EXPRESSION / LABELING:
|
|
spt and ntl translation-blocking morpholinos phenocopy the appropriate single and double mutant phenotypes. (A) Sequences of MOs used to deplete embryos of Spt/Ntl activity. The ntla MO was used in an earlier study (Nasevicius and Ekker, 2000). MOs were injected at a total concentration of 5 ng/nl. To deplete both factors, all four MOs were combined at a concentration of 1.25 ng/nl each and for each single depletion the two appropriate MOs were combined at a concentration of 2.5 ng/nl each. Each embryo was injected with a total of 15 ng to 25 ng of MO. (B-E) Live pictures of 24 hpf embryos following MO injection into one- to four-cell stage embryos. The spt single MO and spt;ntl double MO phenotypes are more necrotic than the corresponding genetic mutants at this stage (Amacher et al., 2002). (F-I) shh expression in the ventral midline is completely abolished posteriorly in 24 hpf embryos depleted of spt and ntl function, and single and double morphant phenotypes mimic those previously demonstrated for genetic mutants (Amacher et al., 2002). Expression of the krox20 hindbrain marker, which labels hindbrain rhombomeres 3 and 5, is included for reference and is not affected in any of the MO-injected embryos. (J-M) Muscle tropomyosin expression is completely abolished in 24 hpf embryos depleted of spt and ntl function, and both single and double morphant phenotypes mimic those previously demonstrated for genetic mutants (Amacher et al., 2002). |
|
Expression of putative T-box gene targets overlaps with ntl and spt expression during gastrulation. Shown are wild-type in situ hybridization patterns during gastrula stages (6-9 hpf) for genes downregulated at least 2.5-fold in embryos depleted of spt and ntl function. Expression patterns obtained from the ZFIN database (Thisse et al., 2001) are indicated. (A) ntl and spt expression patterns at mid-gastrulation. (B) Expression of 18 potential targets, arranged (from top left to bottom right) to indicate the magnitude to which they were affected (fold decrease indicated at the bottom right of each panel). Two additional targets are expressed in patterns that overlap with spt and/or ntl, but are not shown: during gastrulation, knypek (3.8x) is expressed in the axial mesoderm (Machingo et al., 2006) and six4.2 (2.8x) is expressed in the non-axial mesoderm (Kobayashi et al., 2000). An additional nine potential targets that were down-regulated at 2.5-fold or greater are not shown for the following reasons: the gene they identify is not known four genes: AI942866 (8x); AI584322 (6x); BM036916 (4.5x); and BI886648 (4x), their gastrula expression patterns have not been well characterized or described two genes: cxcl12a (2.8x) and itm2c (2.5x), their gastrula expression levels have been reported as low or not detectable two genes: ywhae1 (3.6x) and efemp2 (3x), or they have been described to be ubiquitously expressed during gastrula stages one gene: tuba812 (2.8x). |
|
Spt-GST and Ntl-GST have differential in vitro binding activities. To extend the observation from the SELEX assays that Spt bound a C at position 1 in 25% of the sequenced oligos where as Ntl bound a C at this position in only 2% of the sequenced oligos (Fig. 2), we directly compared the ability of Spt and Ntl to bind TCACACCT versus CCACACCT. Shown are gel mobility shift assays using radiolabeled probes and increasing amounts of bacterially produced Spt-GST and Ntl-GST fusion proteins. Complexes were fractionated on native polyacrylamide gels. Spt-GST bound both probes, but bound TCACACCT probe (A) with higher affinity than the CCACACCT probe (B). As predicted, Ntl-GST also bound the TCACACCT probe (C), but we were unable to detect any Ntl-GST binding to the CCACACCT probe (D). This motif difference has been previously noted between Brachury and Tbx6 in mouse (White and Chapman, 2005), and our data are consistent with previous SELEX experiments with T-box factors, which show that Ntl orthologs do not select sequences with a C at position 1 (Kispert and Herrmann, 1993; Conlon et al., 2001) where as all other tested T-box factors are able to bind oligomers with a C at this position (Conlon et al., 2001; Ghosh et al., 2001; Yagi, 2005; White and Chapman, 2005). |
|
Transgene expression is temporally and spatially regulated like the endogenous gene and is dependent on Spt and Ntl function. (A) Embryos were injected with the tbx6 transgene with or without spt and ntl MOs and the percentage of embryos expressing mCherry at the margin was recorded at various stages of development. (A) The tbx6 transgene is initially activated at shield stage, as has been reported for endogenous tbx6 (Hug et al., 1997) (unbroken line). Injecting spt and ntl MOs eliminated mCherry expression (broken line). Similar experiments examining transgene expression in dld:mCherry stable lines are shown in Fig. 9. (B-E) Double in situ hybridizations for mCherry (in red) and tbx6 (B,C) and deltaD (D,E) (in blue) were performed at 75% epiboly. C and E are higher magnification pictures of the embryos in B and D. In all cases, the majority of mCherry expression occurred in regions that also expressed the endogenous gene, or where the endogenous gene was recently expressed. |