- Title
-
The receptor protein-tyrosine phosphatase, Dep1, acts in arterial/venous cell fate decisions in zebrafish development
- Authors
- Rodriguez, F., Vacaru, A., Overvoorde, J., and den Hertog, J.
- Source
- Full text @ Dev. Biol.
|
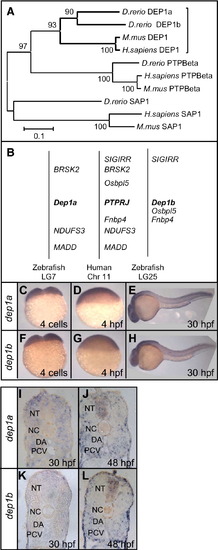
Dep1a and Dep1b are orthologs of human Dep1 and are broadly expressed during early development. (A) Phylogenetic tree of Dep1 and its most closely related family members, PTPβ and SAP1. The PTP domains of zebrafish (D. rerio), human (H. sapiens) and mouse (M. mus) R3 subfamily members were plotted, based on sequence identity. (B) Genomic localization of human Dep1 (Chr. 11), Dep1a (LG7) and Dep1b (LG25) and genes in their immediate surroundings. (C–L) Dep1a- and dep1b-specific riboprobes, encompassing the catalytic domain, were generated and used in whole-mount in situ hybridization. Whole-mount expression of dep1a (C–E) and dep1b (F–H) during early zebrafish development, 4 cell stage (C,F), 4 hpf (D,G) and 30 hpf (E,H) are depicted as well as sections of 30 hpf and 48 hpf embryos (I–L). Relevant structures were labeled: DA, dorsal aorta; NC, notochord, NT, neural tube; PCV, posterior cardinal vein. |
|
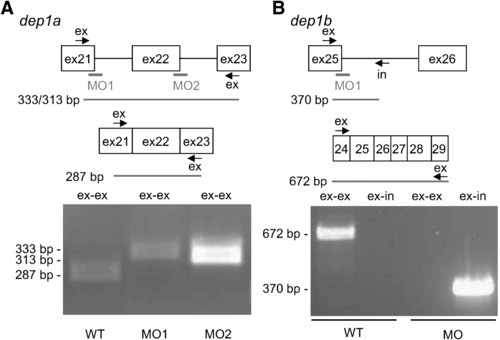
Dep1a- and Dep1b-MOs efficiently blocked splicing. (A) Dep1a-MO1 and Dep1a-MO2 target the 3′ end of exon 21 and exon 22, respectively. Exon-specific primers were designed in exon 21 and exon 23, allowing PCR of spliced RNA (287 bp) and improperly spliced mRNA (333 bp and 313 bp), respectively. RT-PCR was done on non-injected control (WT), Dep1a-MO1 and Dep1a-MO2-injected embryos. The sizes of the obtained PCR fragments on an agarose gel are indicated. (B) The Dep1b-MO targets the 3′ end of exon 25, which encodes sequences corresponding to exon 21 of dep1a (see also Supplementary Fig. S1). Oligos were designed against exon 25 and intron 25, allowing PCR of unspliced dep1b RNA (370 bp) and as a control, exon-specific primers were used (672 bp). RT-PCR was done on RNA, isolated from non-injected control (WT) and Dep1b-MO-injected embryos and the resulting agarose gel is shown with the sizes of the bands indicated. EXPRESSION / LABELING:
|
|
Dep1a-MO induced blood circulation defects, but vasculogenesis appeared normal. O-dianisidine staining of hemoglobin was done on wild type (A) and Dep1a-MO1 (B) injected embryos at 48 hpf. The arrow indicates hemorrhages at the aortic bifurcation and the asterisk blood accumulation around the posterior cardinal vein. Fli1a::egfp1 transgenic zebrafish embryos were injected with Dep1a-MO1 (D) or Dep1a-MO2 (E) at the 1-cell stage and the vasculature was visualized at 30 hpf. A non-injected fli1a::gfp1 embryo served as a control (C). Microangiography was done by injection of rhodamine-conjugated dextran into the heart of embryos. Circulation of the dye was visualized using a fluorescence microscope. Non-injected control (F), Dep1a-MO1 injected (G) and Dep1a-MO2-injected embryos (H) all at 48 hpf are depicted here. (J–K) Magnifications of the area where circulation is blocked (arrow). |
|
Dep1a-MO induced reduction in dorsal aorta marker tbx20. (A) In situ hybridization showing tbx20 expression in wild type zebrafish embryo at 30 hpf and in Dep1a-MO1 (B) or Dep1a-MO2 (C) injected embryos. |
|
Dep1a-MO-induced reduction in tbx20 expression was specific. (A) Classification of the observed defects after Dep1a-MO injection (8 ng/embryo): upper panel, no effect, normal tbx20 expression (white), middle panel, mild effect, patchy tbx20 expression (orange), lower panel, severe effect, no tbx20 expression (red). (B) Quantification of the effects of Dep1a knockdown. The percentages of embryos in the three different classes is given in control embryos (WT) and in Dep1a-MO1 (8 ng) injected embryos. Moreover, Dep1a-MO1 was co-injected with a CMV promoter-driven expression vector for Dep1a (12.5 ng), effectively rescuing the defects. Total numbers of embryos from at least three independent experiments are indicated (n). Exact numbers are given in Supplementary Table S1. (C) Expression vectors for Myc epitope tagged Dep1a and – as a control – for RPTPα were transfected into COS cells. The cells were lysed and whole cell lysates were run on a 7.5% SDS-PAAGE gel. The material on the gel was blotted and the blots were probed with anti-Myc MAb 9E10. An immunoblot is depicted developed with enhanced chemiluminescence and the positions of marker proteins that were co-electrophoresed with the sample are indicated in kDa on the left. |
|
Synergistic effects of Dep1a and Dep1b MOs. Dep1a- and Dep1b-MOs were (co-)injected at the 1-cell stage (4 ng each) and the embryos were fixed at 30 hpf. In situ hybridizations were done using tbx20 specific probes (A–D), a cdh5 (ve-cadherin)-specific probe (E,F) or myoD-specific probes (G,H). (I) Quantification of defects in tbx20 expression, classified as outlined in Fig. 5. The total number of embryos that was analyzed for each condition from three independent experiments is indicated here and depicted in Supplementary Table S2. |
|
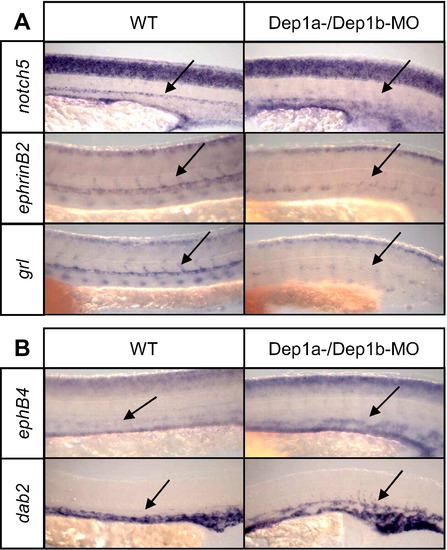
Dep1-MO induced reduction in arterial cell fate. Dep1a- and Dep1b-MOs were co-injected and the embryos were fixed at 30 hpf. Three arterial markers (notch5, ephrinB2 and grl) and two venous markers (ephB4 and dab2) were used for in situ hybridizations. The position of the dorsal aorta is indicated with an arrow. |
Reprinted from Developmental Biology, 324(1), Rodriguez, F., Vacaru, A., Overvoorde, J., and den Hertog, J., The receptor protein-tyrosine phosphatase, Dep1, acts in arterial/venous cell fate decisions in zebrafish development, 122-130, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.