- Title
-
Control of chemokine-guided cell migration by ligand sequestration
- Authors
- Boldajipour, B., Mahabaleshwar, H., Kardash, E., Reichman-Fried, M., Blaser, H., Minina, S., Wilson, D., Xu, Q., and Raz, E.
- Source
- Full text @ Cell
|
Expression Pattern of cxcr7 (A–H) Distribution of cxcr7 mRNA in wild-type embryos during the first 20 hr of development. In situ hybridization using a cxcr7-specific probe shows no staining in four-cell stage embryos (A and B), weak uniform expression at 3.3 hpf (C and D), and enhanced cxcr7 expression in a ring of deep cells at 6 hpf (E and F). At later stages of development (G and H), uniform cxcr7 expression with enhanced expression in mesoderm derivatives and in the nervous system is detected, but no expression is observed at the region where the PGCs are located (encircled domains). (I) Absence of maternally provided cxcr7 mRNA as determined by RT-PCR at the indicated stages. Control reactions are presented in which primers specific for the maternally provided ornithine decarboxylase1 (odc1) mRNA were used. EXPRESSION / LABELING:
|
|
CXCR7 Is Essential for Normal PGC Migration and Is Required in the Somatic Environment of the Embryo (A–C) Reduction of CXCR7 activity leads to aberrant PGC migration as demonstrated by in situ hybridization using a germ cell-specific nanos1 probe. PGCs in control embryos cluster at the region where the gonad develops after 20 hr of development (A). Similar to reduction of CXCR4b activity (B), knockdown of CXCR7 results in a pronounced germ cell migration phenotype (C). (D–F) CXCR7 knockdown does not affect PGC specification. Images of 22 hpf embryos injected with Vasa-GFP-nos1-3′ UTR mRNA are shown. Germ cell-specific mRNA protection and proper localization of the Vasa-GFP fusion protein to germinal granules is observed in embryos injected with control morpholino (D), CXCR4b morpholino (E), and CXCR7 morpholino (F). (G and H) General embryonic patterning and expression of sdf-1a are not affected by CXCR7 knockdown. Two-color in situ hybridization using nanos1 (blue) and sdf-1a probes (red) of embryos injected with control (G) or CXCR7 (H) morpholino is shown. (I) CXCR7 knockdown does not affect PGC number as counted at 12 hpf. n signifies the number of embryos examined. Error bars represent standard error of the mean (SEM). (J–L) The effect of CXCR7 morpholino on PGC migration is reversed by CXCR7 expression. The severe migration phenotype induced by the CXCR7 antisense oligonucleotide (J) is reversed by global expression of RNA encoding CXCR7 (K). A graph demonstrating the dose-dependent rescue of the CXCR7 morpholino-induced phenotype by global expression of CXCR7 (L). For all injections the total amount of injected mRNA was identical (300 pg) by addition of control mRNA (mCherry-F-globin mRNA). The red bar indicates the correct target for the migrating PGCs. (M–Q) PGC migration depends on the activity of CXCR7 in somatic tissues. PGCs expressing DsRedExpress (red) were transplanted into embryos with PGCs expressing EGFP-F (green). Wild-type and CXCR7 knocked-down PGCs arrived at the region of the gonad in wild-type hosts (M and N). A large proportion of wild-type and CXCR7-depleted PGCs does not arrive at the correct target in CXCR7-depleted host embryos (O and P, arrowheads). In (Q), the percent of transplanted PGCs reaching their target after the first day of development isrge proportion of wild-type and CXCR7-depleted PGCs does not arrive at the correct target in CXCR7-depleted host embryos (O a shown. PGCs in CXCR7-depleted hosts embryos show a significant reduction of migration fidelity as compared to PGCs in wild-type hosts (p < 0.001, t test, marked with an asterisk). PGCs deficient for CXCR7 do not show a significant difference in arriving at the target as compared to control PG EXPRESSION / LABELING:
PHENOTYPE:
|
|
CXCR7 Is an SDF-1a Receptor that Promotes the Internalization of the Chemokine (A and B) Subcellular localization of CXCR4b and CXCR7 (green) in somatic cells of the embryo. CXCR4b (green) is predominantly found on the membrane of cells (red label of farnesylated mCherry) (A), while CXCR7 (green) is found on the plasma membrane and intracellularly (B). (C) CXCR7 knockdown increases extracellular SDF-1a levels as judged by internalization of CXCR4b in PGCs. In control embryos (left panel), CXCR4b (green) localizes to the plasma membrane of PGCs (red). CXCR7 knockdown leads to a reduction of CXCR4b on the membrane (middle panel). Membrane localization of CXCR4b in CXCR7 knockdown embryos is restored by SDF-1 knockdown (right panel). (D) SDF-1a is internalized by CXCR7-expressing cells. Somatic cells (red membrane) expressing CXCR7, CXCR4b, or a control protein were transplanted into host embryos that globally expressed SDF-1a-EGFP. Confocal images were taken 1 hr after transplantation. Transplanted cells (red) expressing either control protein or CXCR4b (left and middle panel, respectively) do not show uptake of SDF-1a (green). In contrast, cells expressing CXCR7 showed intracellular accumulations of SDF-1a protein (right panel). (E) SDF-1a and CXCR7 colocalize in vesicular structures. Images were taken 1 hr after transplantation of cells expressing CXCR7-DsRedMonomer into SDF-1a-EGFP-expressing hosts. The inset shows a magnification of the dotted box. (F) SDF-1a accumulates in lysosomes upon CXCR7-mediated internalization. Deconvoluted images were taken 1 hr after transplantation of cells expressing untagged CXCR7 and the lysosomal marker LAMP-1 fused to DsRedMonomer into SDF-1a-EGFP-expressing host embryos. (G–K) CXCR7-expressing cells reduce extracellular SDF-1a levels. In (G) is a graphic illustration of the experiments designed to examine the depletion of SDF-1a from conditioned medium by CXCR7-expressing cells. The conditioned medium was incubated with cells transfected with the different DNA constructs and subsequently transferred to reporter cultures expressing CXCR4b-EGFP. The extent of CXCR4b-EGFP internalization was then determined. In (H), strong CXCR4b internalization is observed in cells exposed to medium treated with control cells. In (I), medium depleted by CXCR4b-expressing cells induced CXCR4b internalization in 87.5% of all reporter cells, compared to control. CXCR4b ), strong CXCR4b internalization is observed in cells exposed to medium treated with control cells. In (I), medium deinternalization was only observed in 56.3% of cells exposed to medium depleted by CXCR7-expressing cells (J). |
|
CXCR7 Controls PGC Polarity by Regulating SDF-1 Levels (A) CXCR7 knockdown reduces the polarity of migrating germ cells. Wild-type PGCs show a typical polarization of the cells with protrusions at the leading edge in the direction of migration (upper panel, arrows). PGCs in CXCR7-depleted embryos exhibit reduced polarity with protrusions extended in opposite directions (lower panel, arrowheads). Cells labeled with EGFP-F. (B) CXCR7 depletion reduces the motility of PGCs in an SDF-1a-dependent manner. The motility of PGCs was followed in time-lapse movies. Error bars represent SEM. Examples for 70 min long migration paths of germ cells are shown. PGCs in CXCR7 morphants exhibit low motility with short tracks that are reminiscent of PGCs migrating in embryos with high-uniform SDF-1a expression (SDF-1a-OEX). Removal of SDF-1 in CXCR7-depleted embryos restores PGC motility to a level that is similar to that in SDF-1-depleted embryos. Similarly, knocking down CXCR4 restores PGC motility in CXCR7 morphants. (C) Reduction of SDF-1a expression suppresses the CXCR7 knockdown phenotype. The migration phenotype of embryos knocked down for CXCR7 (left panel, 66.4% ± 3% ectopic cells per embryo, n = 30 embryos) is suppressed by coinjection of low levels (0.02 pmol) of SDF-1a morpholino (middle panel: SDF-1a-MO, 29.4% ± 3% ectopic cells per embryo, n = 22 embryos; right panel: CXCR7-MO and SDF-1a-MO, 36.0% ± 2% ectopic cells per embryo, n = 63 embryos). |
|
CXCR7 Does Not Activate Major Pathways Downstream to Chemokine Signaling (A) CXCR7 depletion does not alter calcium levels in the cytosol of somatic cells in control and CXCR7-depleted embryos (p > 0.1, t test). n signifies the number of cells examined. Error bars represent SEM. a.u., arbitrary units. (B) CXCR7 knockdown phenotype is not caused by absence of PI3K function. Shown are PGCs expressing DsRed (red) migrating in embryos globally expressing Akt-PH-EGFP. Migration was monitored in CXCR7-depleted embryos and compared with the migration of PGCs in embryos in which PI3K was inhibited. In CXCR7-depleted embryos PGCs display multiple protrusions in opposing directions (upper panel, arrowheads), typical of CXCR7 inhibition. By contrast, PGCs treated with the selective PI3K inhibitor Wortmannin (25 μM) are polar and migrate with the protrusions, forming in the direction of migration (lower panel, arrowheads). Effective inhibition of PI3K function was monitored by the localization of Akt-PH-EGFP. In DMSO-treated embryos (upper panel), the PH domain localizes to the plasma membrane, whereas PI3K inhibition by Wortmannin induces translocation of the sensor to the cytosol (lower panel). Movies of control cells not treated with the drug and cells in CXCR7-depleted embryos treated with Wortmannin are provided in Movies S8 and S9, respectively. (C) Germ cell-specific expression of CXCR7 does not substitute for CXCR4b function. CXCR7 expression does not revert the effect of CXCR4b-deficient fish (gray bars), but rescues CXCR7 morpholino-treated embryos (white bar). n signifies the number of embryos examined. Error bars represent SEM. PHENOTYPE:
|
|
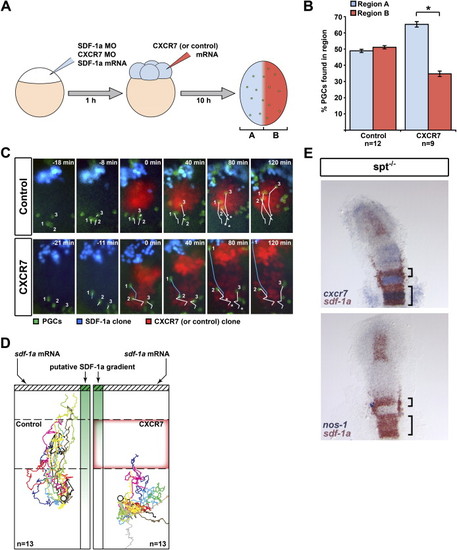
CXCR7 Affects the Direction of Germ Cell Migration In Vivo (A) A schematic representation of the experimental manipulations generating a CXCR7 expression domain (red, Region A) superimposed on uniform SDF-1a expression (blue, Region B). PGCs are depicted in green. (B) In contrast to control experiments, PGCs vacated the CXCR7-expressing B region (p value < 0.001, t test). n signifies the number of embryos examined, and error bars represent SEM. (C) Snapshots of representative time-lapse movies with germ cells (green) migrating toward a transplanted source of SDF-1a (blue) in SDF-1-deficient embryos. A transplant of cells (red) expressing either CXCR7 or control protein was placed at the migration path. In control experiments (upper panel), germ cells (white tracks labeled 1–3) readily traverse the transplant toward the source of SDF-1a. Asterisks denote the starting points. When encountering a CXCR7-expressing transplant (lower panel), the migration toward the SDF-1a source is inhibited (cells 2 and 3). Cells that do not encounter CXCR7-expressing cells on their migration path (cell 1) are not affected (blue track). (D) Multiple migration tracks of germ cells encountering a control transplant (dashed box) or a transplant expressing CXCR7 (red box outline). Tracks have been corrected for morphogenetic movements and were given a common starting coordinate (circle) with the SDF-1a transplant positioned to the top (hatched box). The putative SDF-1a gradient drawn in green. n signifies the number of cells examined. Tracks represent 150 min of PGC migration. (E) Regions expressing sdf-1a fail to attract PGCs if the expression overlaps with that of cxcr7. Two-color in situ hybridization on 13 hpf spt-/- embryos using cxcr7 (blue) and sdf-1a (red) probes (top panel) and nanos1 (blue) and sdf-1a (red) probes (lower panel). EXPRESSION / LABELING:
|
|
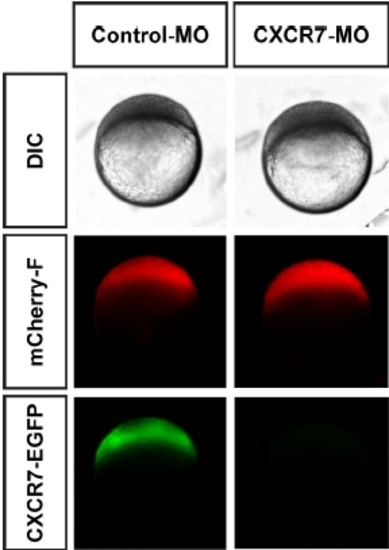
Sequence-specific Knockdown of CXCR7 by CXCR7 Morpholino Injection. Low magnification DIC and epifluorescence images of 4 hpf embryos coinjected with morpholino-sensitive CXCR7-globin mRNA and either control (left panel) or CXCR7 morpholino (right panel). Expression of mCherry-F-globin mRNA serves as control. |
|
SDF-1a Accumulates in Lysosomes Upon CXCR7-Mediated Internalization. Epifluorescent images were taken 1 hour after transplantation of cells expressing untagged CXCR7 and H1M-EGFP-globin (green) into SDF-1a-EGFP (green) expressing host embryos. Lysosomal labelling was obtained by incubation of the embryos in 0.3x Danieau’s buffer containing 70 FM LysoTracker (Invitrogen), starting 1 hour prior to transplantation. |
Reprinted from Cell, 132(3), Boldajipour, B., Mahabaleshwar, H., Kardash, E., Reichman-Fried, M., Blaser, H., Minina, S., Wilson, D., Xu, Q., and Raz, E., Control of chemokine-guided cell migration by ligand sequestration, 463-473, Copyright (2008) with permission from Elsevier. Full text @ Cell