- Title
-
Mutation of DNA primase causes extensive apoptosis of retinal neurons through the activation of DNA damage checkpoint and tumor suppressor p53
- Authors
- Yamaguchi, M., Fujimori-Tonou, N., Yoshimura, Y., Kishi, T., Okamoto, H., and Masai, I.
- Source
- Full text @ Development
|
Retinal neurons undergo extensive apoptosis in the piy mutant. (A) Morphology of wild-type and piy mutant zebrafish embryos at 2 dpf. Both embryos were treated with phenyltiourea to prevent pigmentation. Extensive cell death is observed in the retina and tectum of the piy mutant (arrowheads). (B,C) Plastic sections of wild-type (B) and piy mutant (C) retinas. In the piy mutant retinas, extensive cell death occurs in the central retina, where differentiated neurons are normally located. Retinal stem cells are retained in the CMZ of the piy mutant (asterisks). (D,E) TUNEL of wild-type (D) and piy mutant (E) retinas. In the piy mutant, most cells in the central retina are TUNEL-positive (green), whereas there are only a few TUNEL-positive cells in the CMZ (asterisks). (F,G) BrdU labeling of wild-type (F) and piy mutant (G) retinas. In the piy mutant, retinal stem cells in the CMZ incorporate BrdU (asterisks). PHENOTYPE:
|
|
Apoptosis occurs in differentiating neurons in the piy mutant retina. (A-F) BrdU labeling (A,C,E) and TUNEL (B,D,F) of piy mutant zebrafish retinas. At 32 hpf, many retinal cells incorporate BrdU (A, red) and there are no TUNEL-positive cells (B, red). At 42 hpf, many neurons are produced in the central retina and these express the neuronal marker ath5 (D, green). Apoptotic cells are observed in the ath5-positive central retina, but are rare in the CMZ (D, red). At 72 hpf, dense dead cells are observed in the central retina (F, red) and ath5-positive neurons markedly decrease in number (F, green), suggesting that differentiating neurons undergo apoptosis in the piy mutant. Cells in the CMZ incorporate BrdU (C,E, asterisks), suggesting that retinal stem cells continue to proliferate in the piy mutant. (G,H) In situ hybridization of wild-type (G) and piy mutant (H) retinas with rx1 RNA probe (asterisks). (I,J) Plastic sections of piy mutant retinas treated with DMSO (I) or TSA (J). (K,L) TUNEL (green) of piy mutant retinas treated with DMSO (K) or TSA (L). (M-P) Plastic sections of wild-type (M), smu (N), piy (O) and smu; piy (P) mutant retinas. Although retinal neurogenesis is delayed in the smu mutant, retinal lamination occurs until 3 dpf (N). In the smu; piy double mutant, retinal cell apoptosis is suppressed (P). EXPRESSION / LABELING:
PHENOTYPE:
|
|
piy mutation behaves in a cell-autonomous manner. (A) Transplantation of wild-type zebrafish cells into wild-type recipient retinas. Wild-type donor cells (brown) form retinal columns. (B) Transplantation of wild-type cells into piy mutant recipient retinas. Although most of the recipient cells undergo apoptosis, wild-type donor cells (brown) survive and form retinal columns (arrowheads). (C) Transplantation of piy mutant cells into wild-type recipient retinas. Transplanted mutant cells fail to form retinal columns and instead remain as a very small cell mass (arrowhead). (D) Transplantation of piy mutant cells into wild-type recipient retinas. piy donor mutant cells (red) are TUNEL-positive (green, arrowheads) in the wild-type recipient retina. |
|
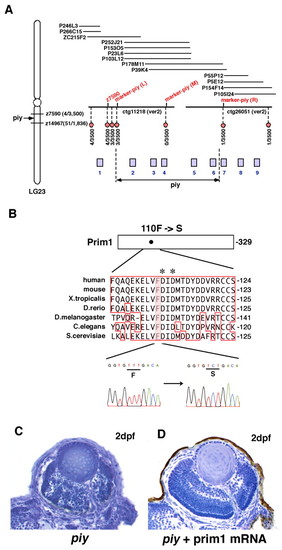
Missense mutation occurs in Prim1 in the piy mutant. (A) Schematic of zebrafish LG23. The piy mutation maps to the genomic region between the SSLP markers z7590 (recombination 4/3500) and z14967 (recombination 51/1836). z7590 maps to the genomic fragment ctg11218, which was shown in the zebrafish genomic database (version 2) released by the Wellcome Trust Sanger Institute. ctg11218 is connected to another genomic fragment, ctg26051, through six overlapping PAC clones. According to the most current version of the zebrafish genomic database, the following nine genes are annotated within this genomic region: 1, zgc:111964 (bHLH transcription factor neuroB); 2, rnd1 (Rho family GTPase 1); 3, prim1 (DNA primase small subunit); 4, zgc:86644 (Tuftelin interacting protein 11); 5, naca (Nascent polypeptide-associated complex alpha); 6, 559664 (EntrezGene) (intracellular membrane-associated calcium-independent phospholipase A2 gamma); 7, 55959 (EntrezGene) (Neurofascin); 8, zgc:65788 (Chitinase); and 9, zgc:123113 (Peptidase). Red circles indicate polymorphic markers that we examined; their recombination rates are shown beneath the red circles. We identified three polymorphic markers, marker-piy (L) (3/3500), marker-piy (M) (0/3500) and marker-piy (R) (1/3500), two of which flanked the piy mutation. Five candidate genes, rnd1, prim1, zgc:86644 (tfip11), naca and 559664 are located in this flanking region. (B) Missense mutation occurred in the prim1 gene in the piy mutant. Amino acid comparison of Prim1 among human (Stadlbauer et al., 1994), mouse (Prussak et al., 1989), Xenopus tropicalis (GenBank accession BC067914), Danio rerio (GenBank accession NM_201448), Drosophila melanogaster (Bakkenist and Cotterill, 1994), Caenorhabditis elegans (GenBank accession NM_06488) and Saccharomyces cerevisiae (Plevani et al., 1987) is shown. Phenylalanine (F) 110 is conserved among these organisms (highlighted). Aspartic acids (D) marked by asterisks indicate the putative active site residues of Prim1 (Augustin et al., 2001). Sequences of the prim1 cDNA in the wild type (left) and piy mutant (right) are shown beneath. The transversion from T to C replaces phenylalanine (F) 110 with serine (S) in the piy mutant. (C,D) piy mutant retinas injected without (C) and with (D) prim1 mRNA. PHENOTYPE:
|
|
The missense mutation F110S does not affect cell proliferation. (A,B) Expression of prim1 mRNA in wild-type zebrafish embryos at the eight cell stage (A) and 2 dpf (B). The maternal expression of prim1 is observed (A). The prim1 gene is predominantly expressed in regions with high cell proliferation rate, such as the eyes and tectum at 2 dpf (B). (C,D) Plastic sections of wild-type retinas labeled with prim1 RNA probe at 2 dpf (C) and 3 dpf (D). The prim1 gene is expressed in the CMZ of the neural retina, suggesting that this gene is expressed in the proliferating cells of the retina. (E,F) Labeling of wild-type (E) and piy mutant (F) retinas with anti-phosphorylated histone H3 antibody (red, arrowheads). All nuclei are counterstained with Sytox Green (green). (G) Percentage of retinal cells that are dividing in wild-type (blue bar) and piy mutant (red bar) retinas at 24 hpf and 32 hpf. (H,I) BrdU labeling (red) of wild-type (H) and piy mutant (I) retinas. (J) Percentage of retinal area that is BrdU-positive in wild-type (blue bar) and piy mutant (red bar) retinas at 24 hpf and 32 hpf. (K,L) Representative traces from FACS analyses after propiduim isodide staining of dissociated cells from wild-type (black line) and piy mutant (green line) heads at 38 hpf (K) and 48 hpf (L). The traces are divided into four regions, M1, M2, M3 and M4, which represent cells with <2N, 2N, 2N-4N and 4N DNA, respectively. (M,N) Bar graph showing summary of FACS results from wild-type sibling and piy homozygous mutant dissociated cell pools at 38 hpf (M) and 48 hpf (N). The piy mutant embryos (light green) contain fewer G1/postmitotic cells and more apoptotic cells than wild type. However, there is no significant difference in the ratio of S/G2/M cells between wild-type and piy mutant embryos. (O) Proliferation rate of yeast pri1-1 mutant cells transformed with wild-type prim1, F110S mutated prim1 and control vector at 37°C. The proliferation rate of the yeast pri1-1 mutant cells transformed with the F110S mutant prim1 (pink) is not significantly different from that of the yeast pri1-1 mutant cells transformed with the wild-type prim1 (blue), whereas the proliferation rate of the yeast pri1-1 mutant cells transformed with the control vector (yellow) is low. |
|
Expression of zebrafish prim1, p53 and chk2 genes. (A-D) Expression of prim1. Lateral views of embryos (A-C) and sectioned retina (D). The expression of the prim1 gene is observed in the entire head at 24 hpf (A) and is prominent in the eyes and tectum at 33 hpf (B). At 48 hpf, piy expression is observed mainly in the proliferative zones of the eye and tectum (C-D). (E-H) The expression pattern of p53 is similar to that of prim1. (I-L) The expression pattern of the zebrafish chk2 gene is similar to that of prim1. |
|
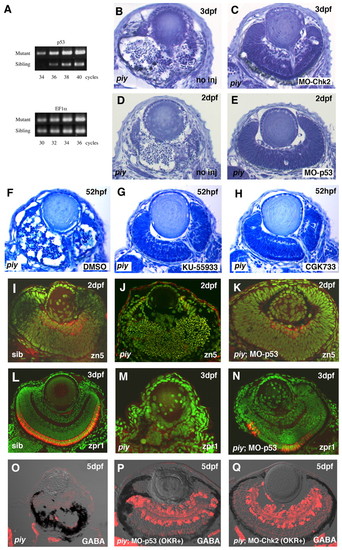
piy-mediated apoptosis depends on the ATM-Chk2-p53 pathway. (A) Quantitative PCR analysis of p53 mRNA in wild-type and piy mutant zebrafish embryos. There is no difference in the amplification of a control gene, ef1α (lower panel), from wild-type and the piy mutant cDNA pools. By contrast, the p53 gene is amplified from the piy mutant cDNA pool by a smaller number of PCR cycles than from the wild-type cDNA pool (upper panel), suggesting that p53 mRNA is more abundant in the piy mutant embryos than in wild-type embryos. (B,C) Plastic sections of piy mutant retinas injected without (B) and with (C) MO-Chk2. (D,E) Plastic sections of piy mutant retinas injected without (D) and with (E) MO-p53. (F-H) Treatment of the piy mutant embryos with DMSO (F), KU55933 (G) or CGK733 (H). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Co-injection of zebrafish embryos with morpholino antisense oligos (MOs) and the hybrid RNA encoding GFP fused to the 5′ untranslated and initial 20-bp coding region of their target genes. (A) Schematic of experimental procedures. The hybrid RNA or a mixture of the hybrid RNA and MOs was injected into one-cell stage embryos. The hybrid RNA activates GFP expression a couple of hours after injection (above), but simultaneous introduction of MOs represses this RNA-mediated GFP expression if MOs specifically inhibit the translation of their target genes (beneath). We examined GFP fluorescence in embryos injected with different combinations of RNAs and Mos: (B) no RNA and no MO; (C) the hybrid RNA encoding GFP fused to the 5′ untranslated and initial 20-bp coding region of the chk1 gene RNA(Chk1:GFP) and no MO; (D) RNA(Chk2:GFP) and no MO; (E) RNA(Chk1:GFP) and MO-Chk1; (F) RNA(Chk2:GFP) and MO-Chk2; (G) RNA(Chk1:GFP) and 5mis-MO-Chk1; (H) RNA(Chk2:GFP) and 5mis-MO-Chk2; (I) RNA(Chk1:GFP) and MO-Chk2; (J) RNA(Chk2:GFP) and MO-Chk1. |
|
Alcian Blue staining of wild-type and piy mutant embryos. (A-D) Ventral (A,B) and lateral (C,D) views of wild-type sibling (A,C) and piy mutant (B,D) embryos. The piy mutant embryos display a similar defect in cartilaginous elements of head skeleton to fla mutant embryos (Plaster et al., 2006). PHENOTYPE:
|

Unillustrated author statements |