- Title
-
Nodal signals mediate interactions between the extra-embryonic and embryonic tissues in zebrafish
- Authors
- Fan, X., Hagos, E.G., Xu, B., Sias, C., Kawakami, K., Burdine, R.D., and Dougan, S.T.
- Source
- Full text @ Dev. Biol.
|
Construction and expression of transient sqtGFP lines. (A) Diagram of the sqt locus in BAC157J11. sqt has three exons (blue rectangles) separated by two introns, 631 bp and 81 bp long, respectively. The start of transcription is indicated (arrow). Key restriction sites and the vector gene encoding chloramphenicol resistance are depicted. The GFP-FRT-neo-FRT cassette is diagrammed below the BAC, showing the site of integration in the first exon. The resulting engineered BAC clone is depicted below. After integration and excision of the gene encoding neomycin resistance, the gfp coding region replaces the sqt first exon. Green rectangle = GFP sequences; yellow rectangle = neomycin resistance gene; red squares = FRT recombination sites; blue rectangle = sqt coding sequences. (B) Diagram of p10sqtGFP following excision from the BAC by gap-repair. HindIII sites are indicated. Red lines indicate the sequences used to generate Tg-SqtapGFP and its derivatives. (C) Fluorescent image of a living, 5 hpf embryo injected with BAC157J11sqtGFP at the one-cell stage. Fluorescence appears throughout the margin, including the YSL, EVL and blastomeres. Perduring GFP expression is also observed in some cells farther from the margin. (D) Low magnification fluorescent image of a living, 7 hpf embryos injected with BAC157J11sqtGFP. Fluorescence is observed in the dorsal forerunners (white arrows). Perdurant expression is also detected in blastomeres and EVL cells farther from the margin. (E) Fluorescent image of a living 5 hpf embryo injected with p9.9sqtGFPi at the one-cell stage. GFP fluorescence is detected around the entire margin, in the blastomeres, EVL and YSL. K = KpnI; X = XhoI; Xb = XbaI; E = EcoR1; N = Not1; H = HindIII. |
|
Spatio-temporal expression of sqtGFP transgenes reflects endogenous sqt expression. (A) Diagram of three constructs used to make stable transgenic lines with the Tol2 transposase. Each line consists of the GFP reporter gene and 1.4 kb of DNA from the sqt genomic region upstream of the transcription start site. Tg-SqtapGFPi contains 600 bp of the sqt first intron inserted downstream of the GFP poly-adenylation signal. Tg-SqtapGFP lacks the entire first intron. Tg-SqtapGFPiΔNRE is identical to Tg-SqtapGFPi, except that it lacks the 68 bp NRE sequence, shown below. The FoxH1 consensus sites are highlighted in red; Smad consensus binding sites are in black bold. Time-course of gfp expression in Tg-SqtapGFPi (B–E), Tg-SqtapGFP (F–I) and Tg-SqtapGFPiΔNRE (J–M). In all lines, gfp mRNA is induced in dorsal blastomeres soon after MBT (B, F, J). At 5 hpf, gfp is expressed in a ring around the entire margin in all lines (C, G, K). The ring of expression in Tg-SqtapGFP (G) and Tg-SqtapGFPiΔNRE (K) is thinner than the ring in Tg-SqtapGFPi. At 6 hpf, gfp expression in Tg-SqtapGFPi (D) and Tg-SqtapGFPiΔNRE (L) is absent from the margin, but expression persists in Tg-SqtapGFP (arrows) (H). At 8hpf, gfp is expressed in the dorsal forerunner cells in all three lines (red arrowheads) (E, I, M). In Tg-SqtapGFP, expression persists in marginal EVL cells throughout gastrulation (arrows) (I). In all three lines, ectopic gfp expression is detected at the midline (black arrowheads) (E, I, M). (N) The FoxH1 consensus binding sites in the sqt intron are conserved in Tetraodon nigroviridis (Tn), Takifugu rubripes (Tr), Xenopus laevis (Xl), and Mus musculus (Mm). |
|
Expression of gfp in the blastomeres depends on Nodal signaling. Distribution of gfp mRNA in sections of 5 hpf Tg-SqtapGFPi (A, E, G), Tg-SqtapGFP (B, F) and Tg-SqtapGFPiΔNRE (H) embryos stained to reveal gfp mRNA. Whole mounts of Tg-SqtapGFPi (C) and Tg-SqtapGFP (D) are also depicted. In all sections, the membrane that separates the YSL from the blastomeres is highlighted in red. (A) In Tg-SqtapGFPi, gfp transcripts are localized in the EVL (arrow), the blastomeres, and the YSL. (B) In Tg-SqtapGFP, gfp is expressed predominantly in the YSL and EVL (arrow). Expression is also observed in rare blastomeres adjacent to the YSL or EVL. (C, E) In response to activation of the Nodal pathway by ubiquitous expression of TARAM-D, gfp is globally expressed in Tg-SqtapGFPi embryos. In section, gfp transcripts are observed in all blastomeres, EVL cells (E, arrows), and the YSL. (D, F) In Tg-SqtapGFP embryos, TARAM-D induces gfp expression throughout the embryo (D), but only in the EVL cells (F, arrow). (G) When Nodal signaling is blocked by treatment with SB-505124 in Tg-SqtapGFPi embryos, gfp expression is reduced, but not eliminated in the YSL and EVL, and is eliminated in the blastomeres. (H) In Tg-SqtapGFPiΔNRE embryos, gfp expression is restricted to the EVL (arrow) and YSL. |
|
Expression of components of the Nodal-signaling pathway. Sections of wild type embryos at 5 hpf (A) or 4.3 hpf (B–D) embryos stained for cyc (A), oep (B), bon/mixer (C) or foxh1 mRNA (D). (A) In 5 hpf embryos, cyc transcripts are distributed in a punctate pattern in marginal blastomeres within 5 rows of the YSL, and are also detected in the YSL (arrows). (B) oep is expressed in all blastomeres and EVL cells, but transcripts are excluded from the YSL. (C) bon/mixer is expressed exclusively in the blastomeres within 3–4 rows of the YSL. bon/mixer mRNA is not detected in the YSL or EVL. (D) foxh1 transcripts are found throughout the embryo, including all blastomeres and the YSL. EXPRESSION / LABELING:
|
|
sqt and cyc have partially overlapping roles in the YSL. Images of live 5 hpf (A), 6 hpf (C, E, G, I), or 24 hpf (B, D, F, H) embryos after targeted co-injection into the YSL of sqtMO and cycMO, or appropriate 5 bp mismatched control MOs. After co-injection of sqt and cyc mismatched MOs, the embryonic shield forms normally (A, arrow) and the body axis is indistinguishable from wild type (B). After co-injection of the sqtMIS MO and cycMO, the embryonic shield forms (C, arrow) and the body axis is normal (D). Embryonic shields do not form in many embryos following injection of the sqtMO and cycMIS MO (G). The majority of these embryos appear normal at 24 hpf (H), but many have reduced cardiac tissue. (I, J) After co-injection of sqtMO and cycMO, the vast majority of embryos lack embryonic shields (G). At 24 hpf, these embryos display severe cyclopia and have reduced notochord due to defects in axial mesoderm patterning (J). Paraxial mesoderm is less affected, as somites form in embryos co-injected with sqtMO and cycMO (J). |
|
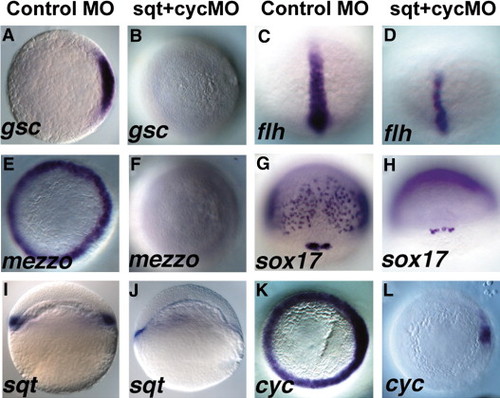
Sqt and Cyc in the YSL are required for endoderm and head mesoderm. Embryos were injected with sqt and cyc mismatch MOs (A, C, E, G, I, K) or sqt and cyc MOs (B, D, F, H, J, L) and processed for in situ hybridization for mesoderm and endoderm marker genes. Control MOs do not affect expression of gsc (A), flh (C), mezzo (E), sox17 (G), sqt (I) or cyc (K). (B) By contrast, gsc expression is eliminated when sqt and cyc MOs are co-injected into the YSL. (D) flh expression is reduced, but not eliminated in these embryos. (F) These embryos also completely lack mezzo expression. (H) sox17 expression is eliminated from the endoderm progenitors, but not from the dorsal forerunners. (J) Extremely low levels of sqt expression are apparent when Sqt and Cyc signals are depleted from the YSL. (L) cyc expression is greatly reduced, except for a small patch of expression in the presumed dorsal blastomeres. In panels A–D, E–F, I–L, dorsal is to the right, when apparent. EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 310(2), Fan, X., Hagos, E.G., Xu, B., Sias, C., Kawakami, K., Burdine, R.D., and Dougan, S.T., Nodal signals mediate interactions between the extra-embryonic and embryonic tissues in zebrafish, 363-378, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.