- Title
-
Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development
- Authors
- Hernandez, R.E., Putzke, A.P., Myers, J.P., Margaretha, L., and Moens, C.B.
- Source
- Full text @ Development
|
Expression of the cyp26 genes in the developing hindbrain. (A-R) Whole-mount in situ hybridizations during the hindbrain patterning period. All embryos are shown as dorsal views. Anterior is to the top in A-I and to the left in J-R. In situ probes are noted in brackets beside the panels, embryonic age is noted in hours post fertilization (hpf). (A) Schematic of an 80% epiboly (8.5 hpf) embryo. The dotted box is the region shown in the flat-mounted embryos in B-E; the arrowhead indicates the advancing margin of the epiblast. During gastrulation, cyp26a1 (B-D,F,G) is expressed in the ectoderm (bracket in B-D) anterior to the domain of RA synthesis indicated by aldh1a2 expression (bracket in E). (C,D) Ectodermal cyp26a1 expression expands in the presence of sub-teratogenic concentrations of RA (C), but is established independent of RA (D). (F,G) cyp26a1 expression recedes anteriorly at the onset of somitogenesis. krox20 (red) is shown in r3 and r5. Arrowhead indicates the posterior limit of cyp26a1 expression. Bracket marks weak cyp26a1expression in the anterior trunk mesoderm. (H,I) aldh1a2 expression during early somitogenesis. Bracket shows expression in trunk mesoderm. (J-R) Dynamic cyp26b1 (J-M) and cyp26c1 (N-R) expression during somitogenesis. krox20 expression in r3 and r5 is in red in J-P; cyp26b1 expression is shown in blue in J-M; and cyp26c1 expression is shown in blue in N-R. In Q,R, cyp26c1 is in red whereas hoxd4 (Q) and vhnf1 (R) are in blue. Insets in Q,R correspond to the dotted boxes. Dotted curve in O indicates the cyp26c1-free domain in ventral r3-r6. Scale bars: 100 μM. Scale bar in B is for B-E; scale bar in F is for F-I; scale bar in J is for J,N,R; scale bar in K is for K,O; scale bar in L is for L,M,P. AN; anterior neurectodermal expression; TB, tailbud expression. EXPRESSION / LABELING:
|
|
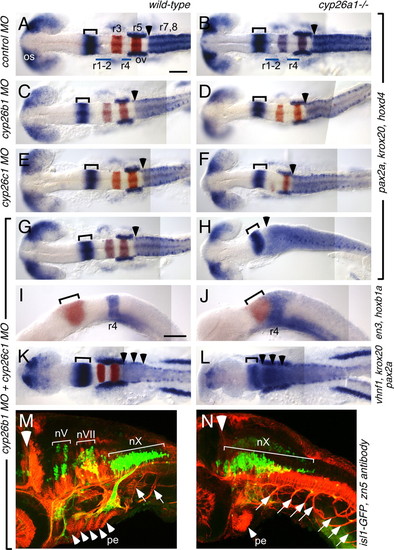
cyp26b1 and cyp26c1 function redundantly with cyp26a1 to pattern the hindbrain. Whole-mount RNA in situ hybridizations at 18 hpf (A-J) and 13 hpf (K,L) and immunostaining at 48 hpf (M,N) in wild-type (left column) and cyp26a1-/- (right column) embryos injected with MOs as shown on the left. (A-H) pax2a (blue) marks the optic stalk (os), posterior midbrain and cerebellum (bracket), and the otic vesicles (ov); whereas hoxd4 (also blue) marks the r7-r8 territory and krox20 (red) marks r3 and r5. MO depletion of Cyp26b1 and/or Cyp26c1 does not affect this pattern in wild-type embryos (C,E,G), but progressively posteriorizes the hindbrain in cyp26a1-/- embryos (D,F,H). Arrowhead marks the r6-r7 boundary, which is shifted to the anterior hindbrain in Cyp26-depleted embryos. (I,J) en3 (red) marks the posterior midbrain and cerebellum (bracket) and hoxb1a (blue) marks r4, which is shifted anteriorly in Cyp26-depleted embryos. (K,L) pax2a (blue) and krox20 (red) are expressed as described above. vhnf1 (also blue) is expressed in the posterior hindbrain up to the r5-r6 boundary (arrowheads) and is also shifted anteriorly in Cyp26-depleted embryos. (M,N) The isl1-GFP transgene (green) marks cranial motor neurons (nV: trigeminal motor neurons in r2 and r3; nVII: facial motor neurons in r4-6; nX: vagal motor neurons in r8) whereas the zn5 antibody (red) marks spinal motor neurons (arrows), pharyngeal arch endoderm (pe, arrowheads mark individual pharyngeal arches) and other structures. The large white arrowhead indicates the mid-hindbrain boundary. In Cyp26-depleted embryos, the motor neurons of the vagus nerve (nX) are expanded anteriorly, as are the spinal motor neurons. Scale bars: 100 μm. Scale bar in A is for A-H,K,L; scale bar in I is for I,J. EXPRESSION / LABELING:
PHENOTYPE:
|
|
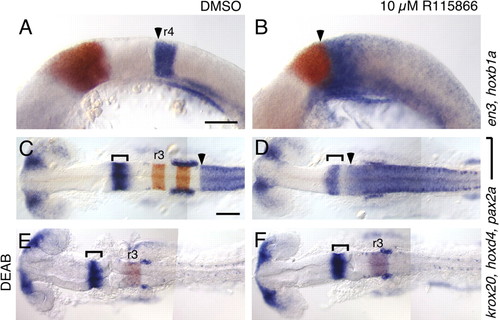
A selective antagonist of Cyp26 enzymes recapitulates the cyp26a1 ; cyp26b1;cyp26c1 phenotype. RNA in situ hybridizations with the markers described in Fig. 2. (A-F) Compared with DMSOtreated controls (A,C,E), treatment with 10 μM R115866 (B,D,F) causes an anterior shift of hoxb1a (arrowhead in A,B) and hoxd4 (arrowhead in C,D) towards the presumptive cerebellum, marked by en3 (red in A,B) and by pax2a (brackets in C-F). This effect of R115866 is reversed by co-treatment with 10 μM DEAB (E,F). Scale bars: 100 μm. Scale bar in A is for A,B; Scale bar in C is for C-F. EXPRESSION / LABELING:
|
|
cyp26a1 protects the hindbrain from exogenous RA. Wild-type (left column) and cyp26a1-/- (right column) embryos treated with DMSO (A,B), 10 μM DEAB (C,D) or 10 μM DEAB + 5 nM RA (E-L). RNA in situ hybridizations use the markers described in Fig. 2, except for I,J, which is a mix of en3 (bracket), krox20 (r3, r5), dlx2 [cranial neural crest (cnc) and forebrain (fb)] and myoD (somites; s). Large bracket in A indicates the r7-r8 region, which is elongated in cyp26a1 mutants (B). (C) In DEAB-treated embryos, posterior rhombomeres (r5-r8) are absent (arrow indicates the absence of high hoxd4 expression characteristic of r7-r8). (D) This phenotype is partially rescued in cyp26a1 mutants, as seen by rescue of r5 but not of r7-r8. (E-L) The DEAB phenotype is fully rescued in wild-type embryos by treatment with 5 nM RA (E,G,I,K) whereas, in cyp26a1 mutants, this low dose of RA causes strong posteriorization of the brain (F,H,J,L). This phenotype resembles that of wild-type embryos treated with 200 nM RA (inset in J). Scale bar: 100 μm. os: optic stalk; e: eye; p: pronephros. EXPRESSION / LABELING:
|
|
Exogenous RA disrupts cyp26b1 and cyp26c1 expression in cyp26a1-/- embryos but not in wild type. cyp26b1 (A-D) and cyp26c1 (E-H) expression (blue) is established normally in wild-type (A,E) and cyp26a1-/- (B,F) embryos at the 6-somite stage (12 hpf). cyp26b1 and cyp26c1 expression is also established normally in wild-type embryos treated with a sub-teratogenic concentration of RA (5 nM; C,G), but not in cyp26a1-/- embryos treated with 5 nM RA (D,H). krox20 expression is shown in red. EXPRESSION / LABELING:
|
|
cyp26a1 protects against teratogenic effects of the RA precursor retinal. Wild-type (A) and cyp26a1-/- (B) embryos injected with 20 pmol retinal at the one-cell stage. Wild-type embryos are only mildly affected by approximately triple the normal levels of retinal, whereas cyp26 mutants are Strongly posteriorized, with hoxd4 expression extending throughout the brain. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
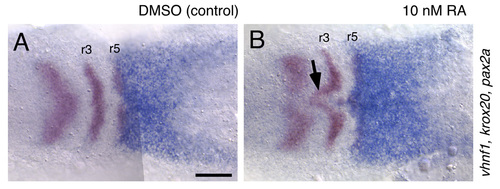
The ventral hindbrain is sensitive to low concentrations of RA. RNA in situ hybridizations showing vhnf1 (blue), and krox20 and pax2a (both in red) expression at the 3-somite stage (11 hpf). In embryos treated with 10 nM RA, which causes very mild, if any, patterning defects, the ventral-most hindbrain is specifically posteriorized, corresponding with the absence of cyp26c1 expression there (arrow in B). Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Effects of RA on cyp26b1 and cyp26c1 expression. RNA in situs at the 3-somite stage showing cyp26bc1 (A-C) and cyp26b1 expressions (D-F). In the presence of 100 nM RA, posterior rhombomeres, including r4, are expanded anteriorly, and the r4 expression of cyp26b1 and cyp26c1 are correspondingly expanded (B,E). In the presence of 10 μM DEAB, which blocks RA synthesis, anterior hindbrain regions, including r2 and r3, are expanded posteriorly, and the r2 and r3 domains of cyp26b1 and cyp26c1 are correspondingly expanded (C,F). Thus, neither gene requires RA for its onset in the hindbrain, although both genes are affected by treatments that alter RA-dependent hindbrain patterning events. EXPRESSION / LABELING:
|
|
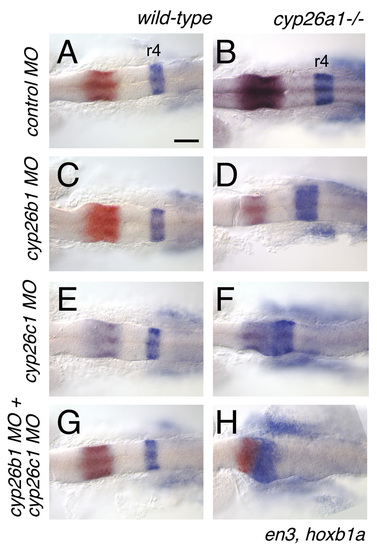
cyp26 genes function redundantly to protect the anterior hindbrain from endogenous RA. cyp26a1 and cyp26c1 are responsible for setting the r3-r4 limit. (A,C,E,G) Knocking-down cyp26b1 and/or c1 has no effect on hindbrain patterning in wild-type embryos. (B) cyp26a1 mutants exhibit a mild expansion of r4. This effect is enhanced by knocking-down cyp26b1 (D), but hoxb1a expression is shifted to the presumptive cerebellum (En-expressing domains) by knocking-down cyp26c1 (F). Embryos depleted for all three cyp26 proteins exhibit a profound anterior shift of hoxb1 expression to the mid-hindbrain domain (H). Scale bar: 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|