- Title
-
Gene Expression Analysis of Zebrafish Heart Regeneration
- Authors
- Lien, C.L., Schebesta, M., Makino, S., Weber, G.J., and Keating, M.T.
- Source
- Full text @ PLoS Biol.
|
Wound Response/Inflammatory Factors, Secreted Molecules, and MMPs Are Upregulated during Zebrafish Heart Regeneration (A) Genes encoding wound response and inflammatory factors, secreted molecules, and MMPs were clustered based on their expression patterns at 3, 7, and 14 dpa. The gene name, gene symbol, and fold change of each gene can be found in Dataset S1. The color chart indicates fold change of expression, and red and green represent increased and decreased expression, respectively. (B) General trend and average fold change of expression of wound response/inflammatory genes, secreted molecules, and MMPs. The wound response/inflammatory genes are expressed early (peak at 3 dpa) during zebrafish heart regeneration. Genes coding for secreted molecules begin to express at 3 dpa and the expression level reach a peak at 7 dpa. The MMPs start to express at 7 dpa and last until 14 dpa. (C) Semiquantitative RT-PCR of a subset of genes coding for secreted molecules, including apoEb (apolipoprotein Eb), vegfc (vascular endothelial growth factor c), granulin A, pdgf-a (platelet-derived growth factor-a; labeled in red), thymosin β4, midkine a, and cxcl12a (chemokine (c-x-c motif) ligand 12a). β-actin was used as a loading control. The RT-PCR results confirmed the temporal expression patterns of these genes as determined by microarray analysis. |
|
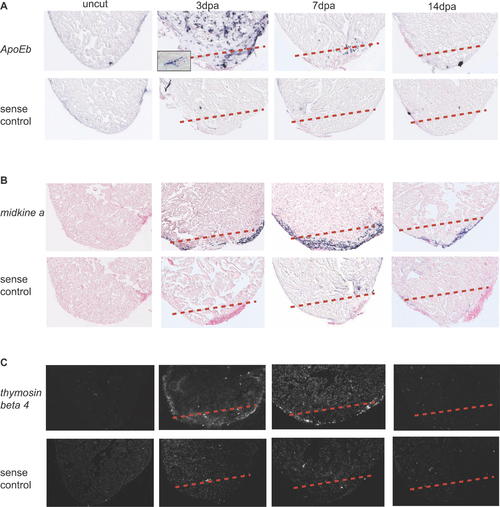
apoEb, midkine a, and thymosin β4 Are Expressed around the Wound Site during Zebrafish Heart Regeneration. Spatial expression patterns of apoEb, midkine a, and thymosin β4 in sham-operated hearts (uncut) and regenerating hearts at 3, 7, and 14 dpa were determined by in situ hybridization. (A) The expression pattern of apoEb was determined using a DIG-labeled antisense probe. apoEb was highly expressed at 3 dpa; its punctate expression suggests that it is expressed by infiltrating macrophages (see the higher magnification image in the inset). (B) Expression level of midkine a was upregulated from 3 dpa, reaches a peak at 7 dpa and lasts until 14 dpa. It appears to be expressed around the wound site in the compact layer of myocardium and the epicardium. (C) The expression pattern of thymosin β4 was determined using a radioactive antisense probe. thymosin β4 appears to be expressed around the wound and surrounding compact myocardium. Sense probes for each gene were used as negative controls. A dashed line marks the amputation plane. EXPRESSION / LABELING:
|
|
pdgf-b Expression Is Upregulated in Regenerating Zebrafish Hearts. (A) RT-PCR analysis of pdgf-b and pdgfr-α. β-actin was used as a loading control. Expression of pdgf-b began to increase at 3 dpa and lasted until 14 dpa during zebrafish heart regeneration. pdgfr-α was expressed in both sham-operated and regenerating hearts; its expression level does not change. (B) In situ hybridization using a radioactive antisense probe showing pdgf-b expression in sham-operated and regenerating hearts at 7 dpa. Expression of pdgf-b was localized to the wound site. A radioactive sense probe was used as a negative control. The dashed red line marks the amputation plane. EXPRESSION / LABELING:
|
|
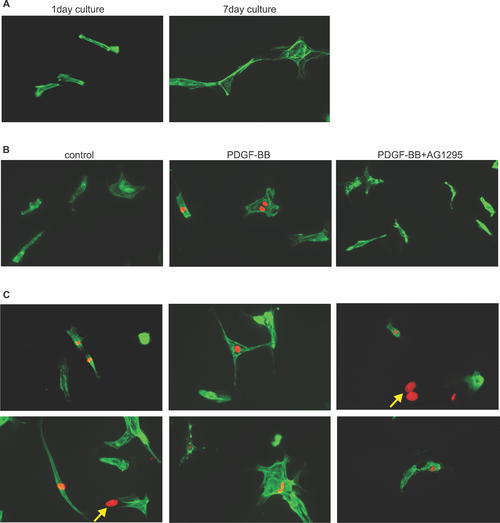
PDGF-B Induces DNA Synthesis in Adult Zebrafish Cardiomyocytes. (A) Adult zebrafish cardiomyocytes were isolated and cultured for 1 or 7 d. The cardiomyocyte population is heterogeneous in size and shape consisting mostly of rod-shape cells. To confirm that the cells are cardiomyocytes, the cells were stained with anti-tropomyosin antibody (green) and anti-MEF2 antibody (unpublished data). After 7 d in culture in 10% FBS, the cardiomyocytes underwent dedifferentiation; the cells lose their rod shape and striation becomes disorganized. (B) DNA synthesis as determined by BrdU incorporation in adult zebrafish cardiomyocytes treated with DMSO (control), PDGF-BB, and PDGF-BB plus the PDGF receptor inhibitor, AG1295. BrdU labeling is shown in red and tropomyosin staining is shown in green. PDGF-BB induced a 2.68-fold increase in DNA synthesis compared to control in adult zebrafish cardiomyocytes (p < 0.05). The effect of PDGF-BB inducing DNA synthesis can be blocked by AG1295. (C) PDGF-BB induced a variety of cardiomyocytes with different size and shape to undergo DNA synthesis. Noncardiomyocytes are marked by yellow arrows. |
|
PDGF Signaling Is Required for Cardiomyocyte Proliferation during Zebrafish Heart Regeneration. After heart surgery, the fish were allowed to recover for one day and were then treated with either the PDGF receptor inhibitor AG1295, DMSO as a negative control, or the MMP inhibitor GM6001 as a specificity control. (A) Heart sections were stained with MEF2 antibody (red) to visualize the cardiomyocyte nuclei and with BrdU antibody (green) to measure DNA synthesis. The BrdU/MEF2 double-positive nuclei (white arrows) are shown in yellow. Many BrdU-positive cells are not cardiomyocytes. PDGF receptor inhibitor treatment resulted in fewer BrdU-positive cardiomyocytes while DMSO and MMP inhibitor treatment did not. The dashed white line marks the amputation plane. (B) Quantification of DNA synthesis in cardiomyocytes during zebrafish heart regeneration. Results were normalized to DMSO-treated control hearts (100%). AG1295 treated hearts had 16% (p < 0.05) fewer BrdU-positive cardiomyocytes compared to DMSO control hearts (Student's t-test), whereas the difference between GM6001-treated and DMSO-treated hearts was statistically insignificant (p > 0.05). Error bars indicate standard error of the mean. |