- Title
-
Zebrafish scl functions independently in hematopoietic and endothelial development
- Authors
- Dooley, K.A., Davidson, A.J., and Zon, L.I.
- Source
- Full text @ Dev. Biol.
|
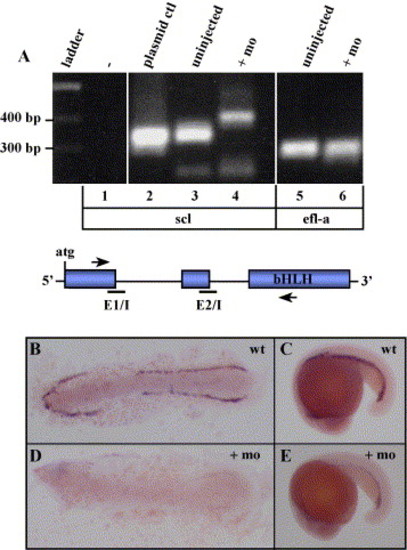
Efficient knockdown of scl by morpholino injection. (A) Reverse-transcriptase (RT)-PCR analysis of uninjected and scl morphant embryos. An aberrant PCR product was observed in the scl morphant (+mo, lane 4) compared to the 325-bp product observed in uninjected control embryos (uninjected, lane 3). An approximate 325 bp product was observed with a pCMV-scl plasmid control (plasmid ctl, lane 2), and no PCR product was observed in the absence of template (-, lane 1). The relative locations of E1/I and E2/I are indicated as black bars in the schematic below, and the relative locations of primers used for RT-PCR are indicated as arrows. As a control, primers designed to amplify ef1-alpha yielded the same sized product from uninjected or scl morphant embryos (lanes 5, 6). B–E) Whole mount RNA in situ hybridization (ISH) for scl in uninjected (B, C) or scl morphant embryos (D, E). In B and D, the yolk was removed and the 10-somite embryos were flat mounted on glass slides for imaging. In C and E, lateral views of the whole embryo at the 20-somite stage are shown. All embryos are oriented with anterior to the left. EXPRESSION / LABELING:
|
|
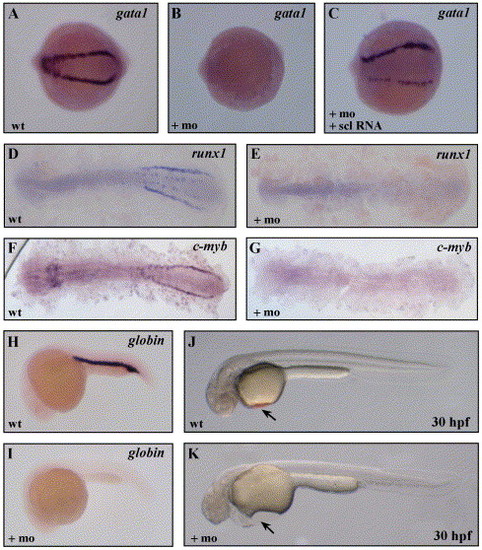
Loss of embryonic erythroid development inscl morphants. (A, B) Loss of gata1 expression as detected by ISH in the scl morphants at the 10-somite stage. (C) Co-injection of 300 pg of scl RNA rescued the gata1 expression. Loss of transcripts for runx1 (D, E) and c-myb (F, G) at the 10 somite stage, and βe3-globin transcripts at 24 hpf (H, I), in scl morphants. (J, K) Absence of circulating erythroid cells in the scl morphant embryos (arrow), in comparison to uninjected control embryos, as visualized under a dissecting microscope. Whole (A–C, H–I) or flat-mounted (D–G) embryos are shown with anterior to the left. EXPRESSION / LABELING:
|
|
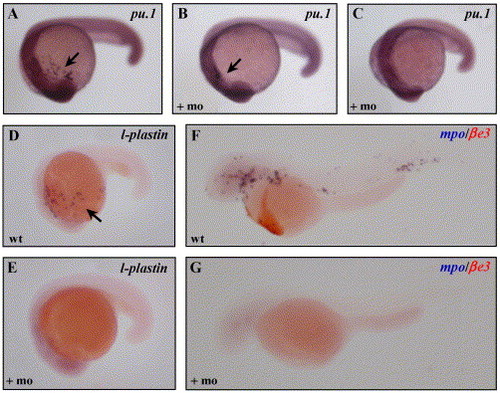
Loss of myeloid development in scl morphants. Transcripts for pu.1 were reduced (B) or absent (C) in the scl morphants, compared to uninjected controls (C), as indicated by the arrows. l-plastin (D, E), and mpo transcripts were not detected in scl morphants. As a control, the mpo ISH was performed as a double with βe3-globin (βe3) staining in red (F, G). |
|
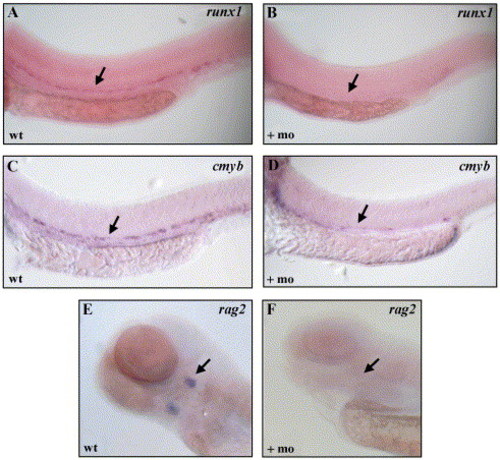
Loss of definitive hematopoietic gene expression in scl morphants. Transcripts for runx1 (A, B), cmyb (C, D), and rag2 (E, F) were absent in scl morphants. In A–D, arrow indicated the region of expression along the ventral wall of the dorsal aorta. In E and F, arrow indicates expression in the thymic rudiment, ventral views. EXPRESSION / LABELING:
|
|
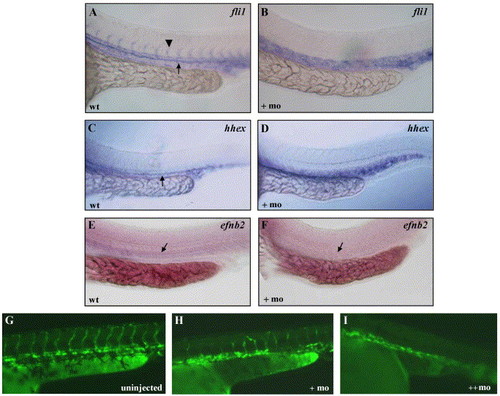
Angiogenesis is disrupted in scl morphants. Transcripts for fli1 (A, B), hhex (C, D), and efnb2 (E, F) in uninjected and scl morphant embryos. In A, the arrowhead indicates the expression in intersomitic vessels, and in A, C, E–F, the arrow indicates expression in the dorsal aorta. G) Uninjected Tg(lmo2:EGFP) embryos, showing fluorescence in the axial and intersomitic vessels and blood cells. In the presence of increasing amounts scl morpholino (H, I), increased disruption of angiogenesis occurred. All images are lateral views of trunk region, anterior to the left. EXPRESSION / LABELING:
|
|
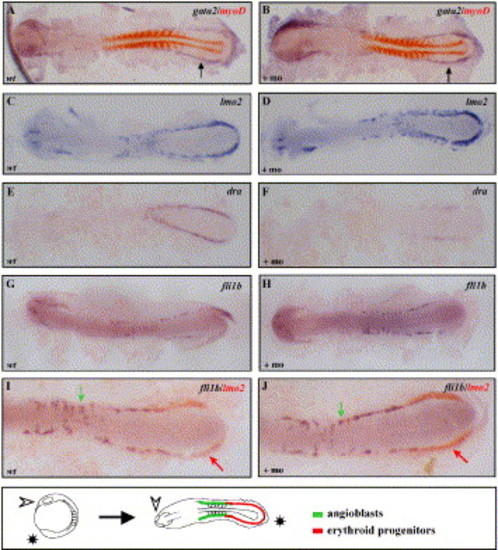
Analysis of early gene expression in the scl morphants. Expression of gata2 (A, B), lmo2 (C, D), dra (E, F) and fli1b (I, J) at the 10- to 12-somite stage in scl morphants and uninjected controls. As an internal control, the gata2 ISH was performed as a double with myoD, a marker of the somitic tissue, detected in red (A, B). (I, J) Double ISH using probes for lmo2 (red arrow) and fli1b (green arrow) in uninjected and scl morphant embryo, showing higher magnification of posterior mesoderm. All embryos were flat mounted and shown with anterior to the left. In D, a schematic illustrates the orientation of embryos and regions of erythroid progenitors and angioblasts. EXPRESSION / LABELING:
|
|
Forced expression of scl RNA in wild-type and spadetail mutants. (A, B) Double ISH for flk1 (blue) or gata1 (red) in uninjected (A) or scl RNA injected (B) embryos. (C, D) myoD transcripts normally present in adaxial cells (black arrow) and somites (blue arrow) are not detected in the scl RNA injected embryos. (E–H) The expression of gata1 (E, F) or flk1 (G, H) in uninjected (E, G) or scl RNA injected (F, H) spt-/- embryos. Brackets indicate anterior head mesoderm; arrow indicates flk1 expression in posterior mesoderm; arrowhead indicates tail bud. All embryos are at the 12-somite stage. EXPRESSION / LABELING:
|
|
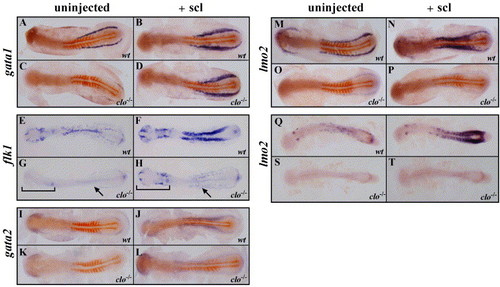
Analysis of hematopoietic and endothelial expansion in clo in response to forced expression of scl. Double ISH (A–D, I–P) using myoD in red and gata1, gata2, or lmo2 in blue, or single ISH using flk1 (E–H) or lmo2 (Q–T), in wild-type and clo-/- embryos injected with scl RNA (+scl) or uninjected controls, as indicated. In G, H, brackets indicate anterior endothelial cells, and arrows indicate posterior lateral mesoderm. In I and J, arrowhead indicates expression of flk1 in the tail bud. Embryos are between the 10- and 12-somite stage (A–P) or the 20 somite stage (Q–T). EXPRESSION / LABELING:
|

Unillustrated author statements EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 277(2), Dooley, K.A., Davidson, A.J., and Zon, L.I., Zebrafish scl functions independently in hematopoietic and endothelial development, 522-536, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.