- Title
-
Wnt1 and wnt10b function redundantly at the zebrafish midbrain-hindbrain boundary
- Authors
- Lekven, A.C., Buckles, G.R., Kostakis, N., and Moon, R.T.
- Source
- Full text @ Dev. Biol.
|
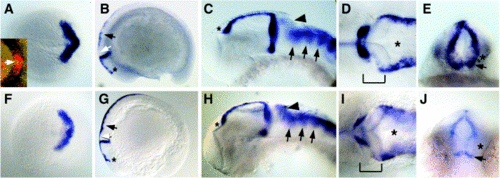
Comparison of wnt1 and wnt10b expression patterns. In situ hybridizations to detect wnt1 (A–E) or wnt10b (F–J) transcripts. In all images except (E) and (J), anterior is to the left. (A, F) Expression at 100% epiboly, dorsal view. Expression of both wnt1 and wnt10b forms a chevron pattern marking the prospective MHB. Inset in (A), double in situ for wnt8b (blue) and wnt1 (red). Arrow indicates region of nonoverlap at the midline. (B, G) Lateral views of embryos at the 16- somite stage. Expression of both genes marks the MHB and dorsal hindbrain/spinal cord (arrows), and both have anterior limits at the prospective epiphysis (asterisks). wnt10b expression is visible in the prospective cerebellum, but wnt1 is not (white arrows). (C–E, H–J) Embryos at approximately 30 hpf. (C, H) Lateral views of heads. wnt1 and wnt10b are both expressed from the epiphysis (asterisks) to the MHB. wnt10b is more strongly expressed at the posterior edge of the cerebellum (arrowheads), but both genes show enrichment in the rhombomeres (arrows). (D, I) Dorsal views of the same embryos as in (C, H). Brackets denote the MHB, and asterisks indicate the hindbrain ventricle. Note that wnt10b expression in the MHB appears slightly weaker than wnt1, but both are expressed on the anterior medial edge of the fold. (E, J) Posterior views of the MHBs of the same embryos as in (C, H). wnt10b expression appears generally weaker, but clear differences are seen in the ventral MHB where both genes are expressed in a stripe above the ventral midline (arrows), but wnt10b appears to be absent immediately above (asterisks). |
|
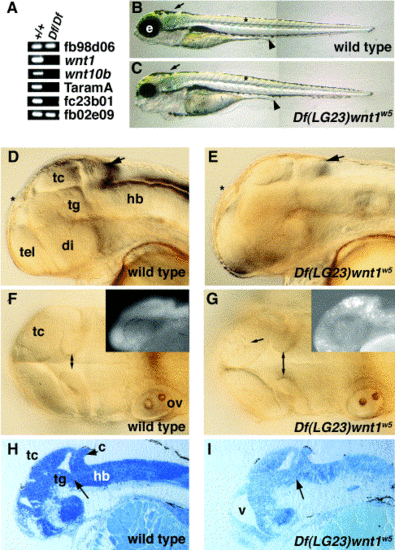
Df(LG23)wnt1w5 is a deficiency for wnt1 and wnt10b. (A) PCR analysis of wild type (+/+) and homozygous mutant DNA (Df/Df). Loci indicated are from the LN54 radiation hybrid panel (see http://zfish.uoregon.edu/ZFIN for mapping panel information). (B, C) Lateral views of 5-dpf embryos, anterior to the left. Melanocytes form normally in Dfw5 embryos (arrows), somites appear normally shaped (asterisks), and the gut tube appears normal (arrowheads), indicating that tissues from all three germ layers are able to form normally. Defects appear most noticeably in the head, as Dfw5 embryos have reduced eyes (e). (D–G) Wild type and Dfw5 embryos at the prim-25 stage (36 hpf). Viewed laterally (D, E), wild type and homozygous mutants are almost indistinguishable (arrows indicate the cerebellum). When viewed dorsally (F, G), an increased distance between the medial edges of the MHB fold is visible in the mutants (double arrows). Additionally, cell death is apparent in the optic tectum (arrow in G). (Insets) Acridine orange staining of 24-hpf embryos. (H, I) Sagittal sections of 48-hpf embryos. Arrow indicates abnormal structure in the Dfw5 tegmentum– hindbrain interface. tc, tectum; tg, tegmentum; hb, hindbrain; tel, telencephalon; di, diencephalon; ov, otic vesicle; v, ventricle. |
|
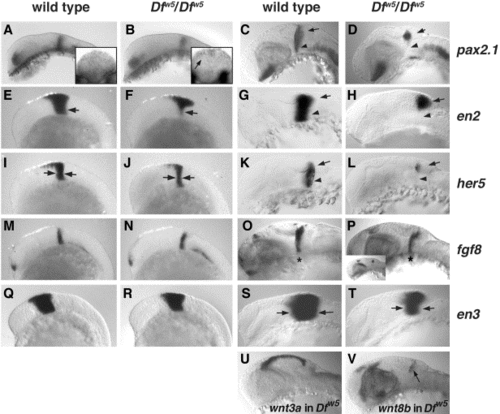
Analysis of MHB markers in Dfw5 embryos. (A, B, E, F, I, J, M, N, Q, R) 12-somite stage embryos. (C, D, O, P) 27 hpf. (G, H, K, L, S, T) 24 hpf. Embryo genotypes are indicated above each column, and probes assayed are indicated to the right of each row. (U, V) 27-hpf Dfw5 embryos; probes are indicated. (All panels) Lateral views of heads, anterior to the left. Reduction in pax2.1 in Dfw5 is first noticeable as a weaker staining at the MHB at the 12-somite stage (B, arrow). (A, B) Insets: Posterior views of MHB pax2.1 expression. Arrow in (B) inset indicates dorsal pax2.1 expression. At 27 hpf, ventral expression of pax2.1 is absent (arrowheads in C, D) leaving only a spot of pax2.1 expression between the cerebellum (arrows in C, D, G, H, K, L) and optic tectum. The reduction in en2 is more clearly seen as a loss of ventral expression at 12 somites (compare arrows in E, F), but at 24 hpf, en2 expression mirrors that of pax2.1 (compare G, H with C, D). The reduction of her5 in 12-somite-stage Dfw5 embryos is visible as a reduction in the entire expression domain (arrows in J indicate thinner band of her5 expression). At 24 hpf, loss of ventral her5 in Dfw5 embryos is evident (L, arrowhead). In contrast, fgf8 expression frequently appears normal in Dfw5 embryos at 12 somites (M, N) and 24 hpf (O, P; asterisk indicated MHB expression domain). Similarly, en3 expression in 12-somite Dfw5 embryos is indistinguishable from wild type (Q, R), but an overall reduction in expression is clear at 24 hpf (S, T; compare width of band between arrows). (U) Expression of wnt3a is indistinguishable between wild type and Dfw5. (V) Expression of wnt8b is indistinguishable between wild type and Dfw5. |
|
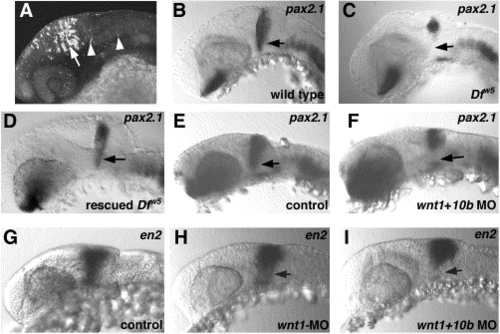
Functional redundancy of wnt1 and wnt10b. (A) Confocal image of GFP expression from PAC 176DG16. Lateral view of head of 24-hpf embryo to illustrate strong expression in the tectum (arrow). Expressing cells are also found in the MHB and hindbrain (arrowheads). (B–I) Probe used is indicated in upper right of each panel; genotype is in lower right. (B–D) Rescue of pax2.1 expression in Dfw5 embryos by PAC injection. (B) Wild type embryo. (C) Dfw5 homozygote to show loss of pax2.1 in ventral MHB (arrow). (D) Dfw5 homozygote with restored ventral pax2.1 expression (arrow) due to injection of PAC176ΔG16. (E–I) Phenotypes induced by wnt1 and wnt10b morpholinos. (E) Control uninjected embryo. pax2.1 expression observed in embryos injected with either wnt1-MO or wnt10b-MO is indistinguishable from this example. Note that MHB pax2.1 expression appears normal (arrow). (F) Embryo coinjected with both wnt1-MO and wnt10b-MO. Note loss of ventral pax2.1 expression as seen in Dfw5 homozygotes (compare with embryo in C). (G) en2 expression in control uninjected 24-hpf embryo. Embryos injected with wnt10b-MO are indistinguishable from this example. (H) Weak reduction in en2 expression seen in ∼30% of embryos injected with wnt1-MO. Note reduction localized to the ventral MHB (arrow). (I) Stronger reduction in en2 expression upon wnt1-MO or wnt1 + wnt10b- MO injection. Compare this phenotype with the Dfw5 embryo in Fig. 4H. |
|
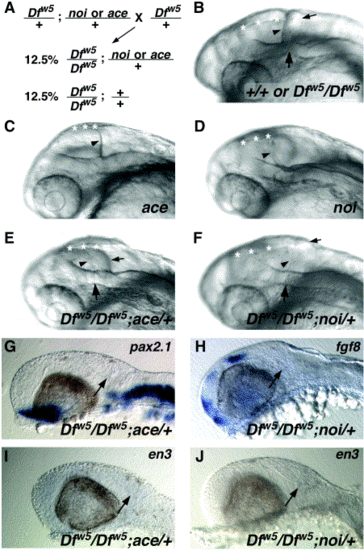
ace and noi are dominant enhancers of Dfw5. (A) Schematic diagram of the cross performed to generate embryos homozygous for Dfw5 and heterozygous for ace or noi. (B–F) Oblique views of heads of live 27-hpf embryos. Genotypes are indicated in the lower right corners. (B) In wild type or Dfw5 homozygotes, the fold is visible and creates a clear separation between the posterior edge of the tectum (arrowhead) and the cerebellum (small arrow). The midline of the fold (large arrow) is visible as an indentation in the lateral hindbrain wall. Asterisks in all panels: dorsal midline of the midbrain. (C) ace mutants fail to form a MHB fold and do not form a cerebellum. Rather, the posterior edge of the optic tectum is in a position corresponding to the position of the cerebellum in wild types (arrowhead). (D) noi mutants also fail to form the MHB fold and cerebellum, but also have defects in the formation of the midbrain. Thus, the tectum appears much smaller than in ace mutants and also has a turbid appearance (not visible). Dfw5 mutants enhanced by ace (E) or noi (F) alleles appear similar in that the separation between the tectum and cerebellum is less distinct due to the absence of a definitive MHB fold. Arrowheads indicate the posterior edge of the tectum, and small arrows indicate the position of the cerebellum. A very slight bump in the hindbrain wall is visible (large arrow). (G–J) Loss of MHB gene expression in Dfw5 embryos enhanced by ace (G, I) or noi (H, J). Probe used is indicated in the upper right corner. Note the absence of expression of each gene in the MHB region (arrows) in comparison to Dfw5 homozygotes (Fig. 4). |

Unillustrated author statements PHENOTYPE:
|
Reprinted from Developmental Biology, 254(2), Lekven, A.C., Buckles, G.R., Kostakis, N., and Moon, R.T., Wnt1 and wnt10b function redundantly at the zebrafish midbrain-hindbrain boundary, 172-187, Copyright (2003) with permission from Elsevier. Full text @ Dev. Biol.