- Title
-
Migration of zebrafish spinal motor nerves into the periphery requires multiple myotome-derived cues
- Authors
- Zeller, J., Schneider, V., Malayaman, S., Higashijima, S., Okamoto, H., Gui, J., Lin, S., and Granato, M.
- Source
- Full text @ Dev. Biol.
|
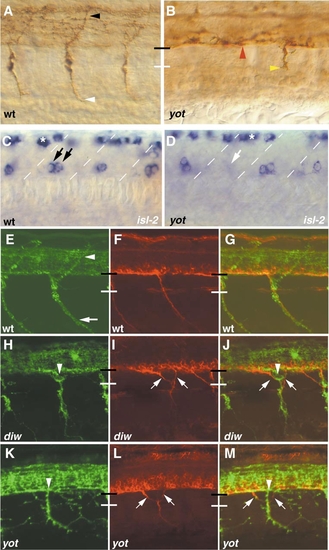
you-too/gli2 embryos lack adaxial cells and display pathfinding defects. Cross-section of wild-type (A) and you-too/gli2 (B) embryos at 28 hpf stained for pax9a expression. In wild-type and you-too/gli2 mutants, pax9a-positive sclerotome cells migrate dorsally, between the notochord and the myotome, encircling the notochord (arrow) and the spinal cord (arrowhead). Note that the horizontal myoseptum is missing in you-too/gli2 embryos, and the somites therefore appear wider. (C, D) Cross-section of a 24-hpf wild-type sibling and you-too/gli2 mutant embryos stained with F59 (red) and F310 (green) antibody. (A) In wild-type siblings F310 (green) stains the fast-twitch muscle cells, while F59 (red) stains the lateral adaxial cell derived slow-twitch muscle cells. (B) In you-too/gli2 mutant embryos, fast-twitch muscle is present (green), while adaxial cell derived slow-twitch muscle is absent. Lateral views of wild-type (E) and you-too/gli2 (F) embryos at 52 hpf stained with zn-5 antibody. Note the expression of zn-5 in the somata of secondary motor neurons (asterisks). In wild-type embryos, (E) secondary motor axons have completed migration along the common path (between red and white line), and extend along their cell-type-specific paths into the dorsal (white arrowhead), medial (yellow arrowhead), and ventral myotome (red arrowhead), respectively. In you-too/gli2 mutant embryos (F), secondary motor axons extend normally inside the spinal cord (white arrows) but often stall at the segmental exit point, where their growth cones accumulated (red arrows), instead of exiting into the periphery. |
|
Pathfinding defects in diwanka mutant embryos. In 52-hpf wild-type embryos (A), secondary motor axons have completed migration along the common path (between red and white line) and extend on their cell-type-specific paths to the dorsal (white arrowhead), medial (yellow arrowhead), and ventral myotome (red arrowhead), respectively. In diwanka mutant embryos (B), zn-5 staining reveals multiple pathfinding defects. Motor axons exited the spinal cord within two independent segmental nerves (asterisk), ventral nerves appeared thinner and less fasciculated and often projected aberrantly into neighboring hemisegments (red arrow). Rostral nerves were absent or less fasiculated (yellow arrow), while projections into the dorsal myotome were absent in all hemisegments. In wild-type embryos, TG(GATA2:GFP)-expressing motor nerves have completed migration along the common path and extended on their cell-type specific paths into the ventral myotome (C). The ventral nerve has formed secondary and tertiary branches that do not cross segmental boundaries. The white line indicates the distal end of the common path. In diwanka mutant embryos (D), TG(GATA2:GFP)-expressing secondary motor axons exit the cord and project though multiple segmental nerves (white arrow), appear thinner and less fasciculated and invade adjacent somites (yellow arrows). In wild-type embryos carrying the TG(ISL1:GFP) transgene (E), labeled motor axons can be detected within the spinal cord extending towards the segmental exit point (red arrowhead), along the common path (white arrowhead), and along their cell-type-specific path into the dorsal myotome (yellow arrowhead). In diwanka mutant embryos expressing TG(ISL1:GFP) (F), secondary motor axons extend inside the spinal cord (red arrowhead) but completely fail to enter the common path. Instead their growth cones stall and accumulate at the segmental exit point (yellow arrows). Schematic summary of wild-type trajectories (G) and diwanka pathfinding defects (H) using TG(GATA2:GFP) (green) and TG(ISL1:GFP) transgenic lines. |
|
The diwanka gene acts cell-nonautonomously to specifically control the migration of motor axons. embryos. In diwanka mutant embryos (B), zn-5 staining of retinal ganglion cells (asterisk) and the optic nerve appear indistinguishable from wild-type embryos at 72 hpf (A). Engrailed (4D9) staining of wild-type (C) and diwanka mutant (D) embryos at 26 hpf. In wild-type and mutant embryos, Engrailed-positive muscle pioneers are located in the anterior portion of the somites. Dashed lines indicate somitic boundaries. (E, F) Chimera analysis of diwanka mutant embryos with a large secondary motor neuron clone. (E) Confocal image of the zn5 stained chimeric diwanka embryo reveals aberrant motor trajectories (arrows and arrowheads). Same embryo and confocal section, now including the rhodamine signal to visualize wild-type derived cells in red. In a diwanka mutant host embryo, wild-type derived motor axons display aberrant, diwanka-like pathfinding defects (arrows, arrowheads and asterisk). (G, H) GFP-positive secondary motor neurons from diwanka mutant embryos expressing TG(GATA2:GFP) were transplanted into wild-type hosts. (G) GFP-positive diwanka motor axons extend along wild-type trajectories, demonstrating that diwanka acts cell-nonautonomously. (H) Same embryo and confocal section, now including the rhodamine signal to visualize all wild-type-derived cells in red. White asterisks indicate motor neuron soma, and white arrows point to their axonal trajectories. |
|
In diwanka and you-too/gli2 mutants, pathfinding errors of secondary motoneurons occur independent of primary motor axons. Lateral views of a wild-type (A) and a you-too/gli2 mutant (B) embryo at 23 hpf stained with znp-1 antibody. (A) In each wild-type hemisegment, a CaP (white arrowhead) and a MiP axon (black arrowhead) are present. (B) In you-too/gli2 embryos, primary motor axons migrate within the spinal cord (red arrowhead), and in only 20% of the hemisegments, motor axons exit the spinal cord but stall at the distal end of the common path (yellow arrowhead). In over 80% of the hemisegments, primary motor axons completely fail to exit into the periphery (n = 100). (C, D) In the ventral spinal cord islet-2 labels CaP and VaP primary motor neurons (black arrows; VaP neurons are present in only 50% of wild-type hemisegments). In you-too/gli2 embryos (D), there is a slight reduction in the number of primary motor neurons (white arrow). Asterisks indicate islet-2-positive sensory neurons in the dorsal spinal cord. (E-M) Confocal analysis of double labeled motor neurons using znp-1 (green, E, H, K) and zn-5 (red, F, I, L) in 38-hpf wild-type (E–G), diwanka (H–J), and you-too/gli2 embryos (K–M). (G, J, M) Both channels. (E–G) In wild-type embryos, znp-1 labels primary and secondary motor axons extending along the common path (between black and white bars), and along the ventral path (white arrow). Note that the dorsal path (white arrowhead) is znp-1-positive, but zn-5-negative, indicating that at 38 hpf, fasciculated secondary motor axons have not extended along the dorsal path. (H–J) In diwanka mutant embryos, zn5-positive motor axons exit ectopically from the spinal cord (arrows) ignoring the path of pioneering znp-1-positive primary motor axons (arrowhead). (K–M) you-too/gli2 embryos display similar phenotypes, demonstrating that, unlike wild-type axons, mutant secondary motor axons (zn5-positive, in red) can ignore pioneering primary motor axons. |
|
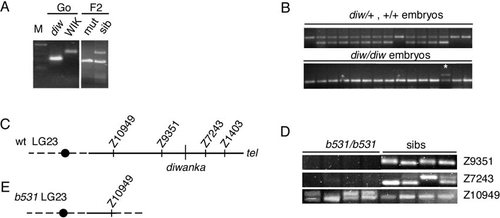
The diwanka locus maps on LG 23 and is deleted in b531 mutant embryos. (A) PCR amplification using marker z1403 and DNA isolated from the G0 diwanka Tü fish, the G0 WIK fish, pooled F2 mutants, and pooled F2 siblings. The Tü-specific PCR fragment is shorter than the WIK-specific fragment. Amplification with z1403 in the F2 sibling pool DNA results in a Tü-specific and a WIK-specific fragment, while in DNA from pooled mutant only the Tü-specific fragment is present (higher mobility), indicating linkage between z1403 and the diwanka mutation. (B) PCR amplification using marker z1403 and DNA from individual F2 sibling embryos (top). (Bottom) PCR amplification using marker z1403 and DNA from individual F2 diwanka mutant embryos. In a recombinant embryo, a Tü-specific and a WIK-specific fragment are present (*). (C) Schematic representation of LG 23 around the diwanka locus. A black circle indicates the centromere. (D) PCR amplification of SSLP markers from DNA of four b531/b531 mutant embryos and four sibling embryos. In sibling embryos (sibs), all tested markers are present. In the four b531/b531 embryos, marker z10949 is present, while markers z9351 and z7243 are absent. (E) Schematic representation of LG 23 in b531/b531 embryos. In these embryos, LG 23 breaks between z10949 and z9351, deleting the remaining chromosomal arm, including the diwanka locus. |
Reprinted from Developmental Biology, 252(2), Zeller, J., Schneider, V., Malayaman, S., Higashijima, S., Okamoto, H., Gui, J., Lin, S., and Granato, M., Migration of zebrafish spinal motor nerves into the periphery requires multiple myotome-derived cues, 241-256, Copyright (2002) with permission from Elsevier. Full text @ Dev. Biol.