The Zebrafish Science Monitor Vol 3(5)
HALF- TETRAD ANALYSIS IN ZEBRAFISH: MAPPING THE ROS MUTATION AND THE CENTROMERE OF LINKAGE GROUP IBy S.L. Johnson, D. Africa, S. Horne, and J.H. Postlethwait, Institute of Neuroscience, 1254 University of Oregon, Eugene, OR 97403-1254, USA
(Genetics 139:1727- 1735,1995)
Analysis of meiotic tetrads is routinely used to determine genetic linkage in various fungi. Here we apply tetrad analysis to the study of genetic linkage in a vertebrate. The half- tetrad genotypes of gynogenetic diploid zebrafish produced by early pressure (EP) treatment were used to investigate the linkage relationships of two recessive pigment pattern mutations, leopard (leo) and rose (ros). The results showed that ros is tightly linked to its centromere and leo maps 31 cM from its centromere. Analysis of half- tetrads segregating for ros and leo in repulsion revealed no homozygous ros individuals among 32 homozygous leo half- tetrads - - i.e., a parental ditype (PD) to nonparental ditype (NPD) ratio of 32:0. This result shows that ros is linked to leo, a mutation previously mapped to Linkage Group I. Investigation of PCR- based DNA polymorphisms on Linkage Group I confirmed the location of ros near the centromere of this linkage group. We propose an efficient, generally useful method to assign new mutations to a linkage group in zebrafish by determining which of 25 polymerase chain reaction (PCR)- based centromere markers shows a significant excess of PD to NPD in half- tetrad fish.
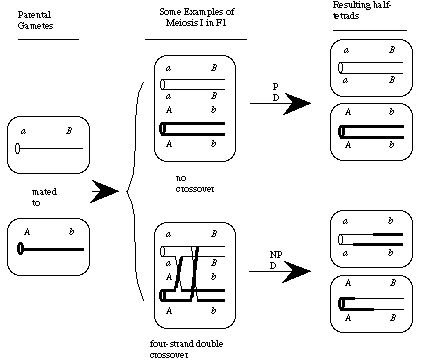 Mutations can be efficiently mapped by half-tetrad analysis in zebrafish. Only no-crossover and four-strand double crossover events between a centromere (a) and a mutant locus (b) can lead to homozygosity at the mutant locus. Because four-strand double-crossovers will be infrequent, linkage group determination is achieved by surveys of centromere markers in selected homozygous mutant half-tetrads. Markers near the linked centromere will show an excess of parental di-type (PD) half-tetrads over nonparental di-type (NPD) half-tetrads, while markers near unlinked centromeres will show equal numbers of NDP and PD half-tetrads among the homozygous half-tetrads. Adapted from Figure 1. Johnson et al. Genetics 139: 1727-1735.
Mutations can be efficiently mapped by half-tetrad analysis in zebrafish. Only no-crossover and four-strand double crossover events between a centromere (a) and a mutant locus (b) can lead to homozygosity at the mutant locus. Because four-strand double-crossovers will be infrequent, linkage group determination is achieved by surveys of centromere markers in selected homozygous mutant half-tetrads. Markers near the linked centromere will show an excess of parental di-type (PD) half-tetrads over nonparental di-type (NPD) half-tetrads, while markers near unlinked centromeres will show equal numbers of NDP and PD half-tetrads among the homozygous half-tetrads. Adapted from Figure 1. Johnson et al. Genetics 139: 1727-1735.
Zebrafish Science Monitor Vol 3(5)
Return to Contents
