- Title
-
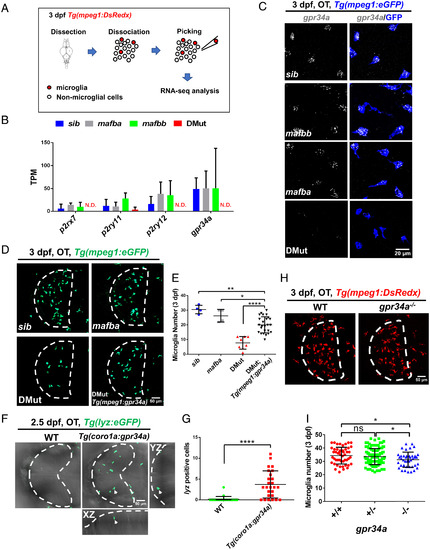
Mafba and Mafbb regulate microglial colonization of zebrafish brain via controlling chemotaxis receptor expression
- Authors
- Lou, L., Yu, T., Dai, Y., Zhao, S., Feng, S., Xu, J., Wen, Z.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|