- Title
-
A Combined Human in Silico and CRISPR/Cas9-Mediated in Vivo Zebrafish Based Approach to Provide Phenotypic Data for Supporting Early Target Validation
- Authors
- Winter, M.J., Ono, Y., Ball, J.S., Walentinsson, A., Michaelsson, E., Tochwin, A., Scholpp, S., Tyler, C.R., Rees, S., Hetheridge, M.J., Bohlooly-Y, M.
- Source
- Full text @ Front Pharmacol
|
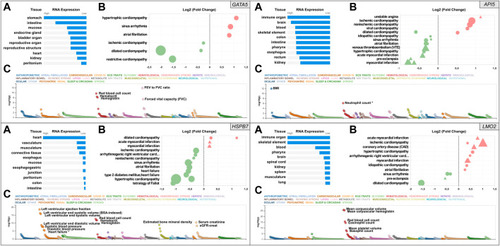
Snapshot of the results from the |
|
Mutation efficiency of the gRNAs for each candidate gene. Data for |
|
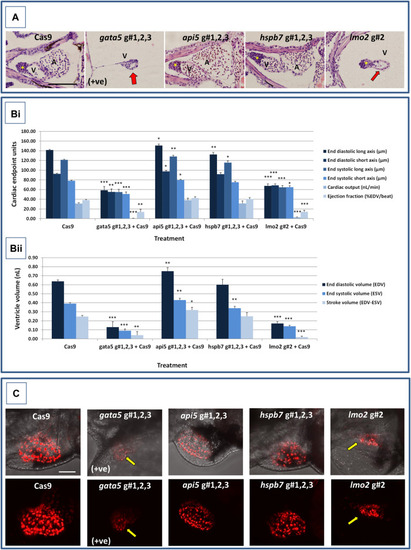
Results of the morphological analysis of 4 dpf PHENOTYPE:
|
|
Results of the cardiac pathological analysis of 4 dpf PHENOTYPE:
|
|
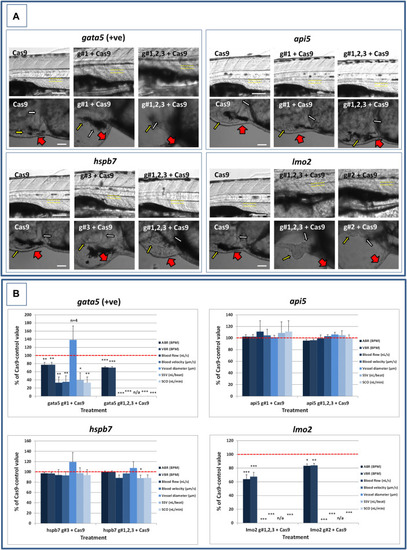
Results of the analysis of cardiovascular function in 4 dpf PHENOTYPE:
|