- Title
-
Identification and Tissue-Specific Characterization of Novel SHOX-Regulated Genes in Zebrafish Highlights SOX Family Members Among Other Genes
- Authors
- Hoffmann, S., Roeth, R., Diebold, S., Gogel, J., Hassel, D., Just, S., Rappold, G.A.
- Source
- Full text @ Front Genet

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
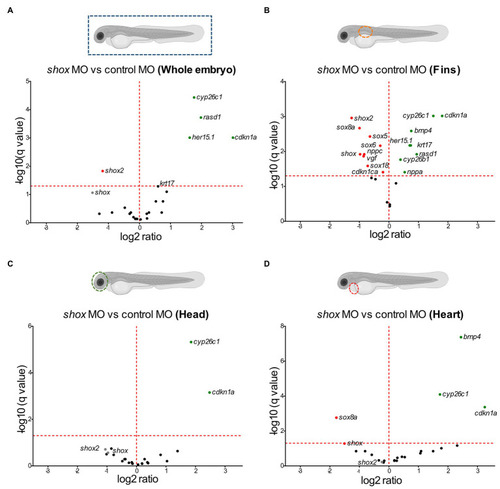
Microarray analysis of |
|
Tissue-specific validation of putative Shox target genes in zebrafish embryos after |
|
Validation of putative Shox target genes in zebrafish fins. Tissue-specific analysis of all selected shox targets upon morpholino-mediated |
|
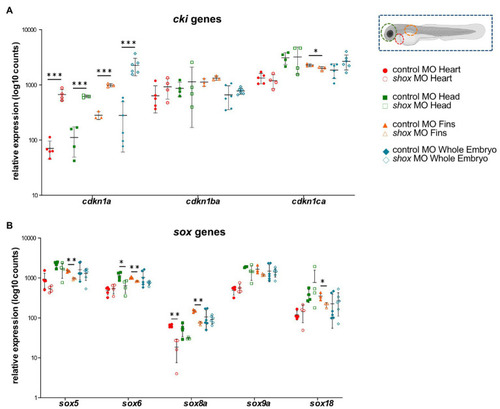
Tissue-specific analysis of gene families upon morpholino-mediated |
|
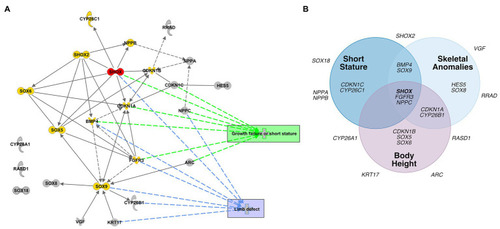
Database analyses of SHOX-regulated genes. |