- Title
-
Reconstruction scheme for excitatory and inhibitory dynamics with quenched disorder: application to zebrafish imaging
- Authors
- Chicchi, L., Cecchini, G., Adam, I., de Vito, G., Livi, R., Pavone, F.S., Silvestri, L., Turrini, L., Vanzi, F., Fanelli, D.
- Source
- Full text @ J Comput Neurosci
|
|
|
Main steps of experimental data elaboration. Every layer of the imaged 3D zebrafish brain is spatially downsampled, as shown in panel |
|
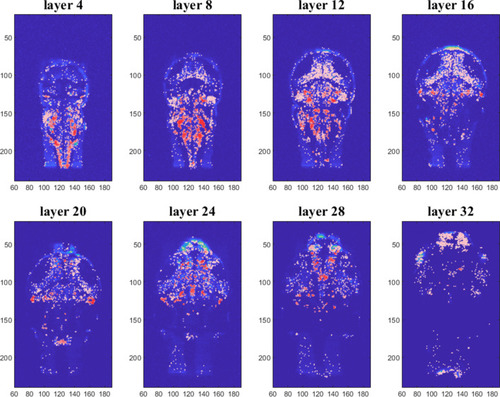
Detected neurons for eight different layers of the zebrafish brain. Colours represent the average cross-correlation of each neuron with all the others selected neurons of the brain |
|
|