- Title
-
In-silico definition of the Drosophila melanogaster matrisome
- Authors
- Davis, M.N., Horne-Badovinac, S., Naba, A.
- Source
- Full text @ Matrix Biol Plus
|
Bioinformatic workflow to define the in-silico matrisome of Drosophila melanogaster. The databases FlyBase, DIOPT, and Ensembl were interrogated with the full list of human and mouse matrisome and matrisome-associated gene symbols. Selected InterPro domains, including domains characteristic of collagens, proteoglycans, ECM-affiliated proteins, and cuticle-binding proteins (see Supplementary Table 4) were used to identify ECM-domain-containing proteins in the reference proteome. The Gene Ontology annotations related to the ECM were then used to identify previously-annotated ECM components. Finally, selected published literature using proteomic and/or bioinformatic methods, as well as reviews on the subject, were searched to identify ECM proteins not identified by the orthology-based or protein-sequencing methods. These data were combined and manually curated to generate the first complete Drosophila matrisome. Hs, Homo sapiens; Mm, Mus musculus; Dm Drosophila melanogaster. |
|
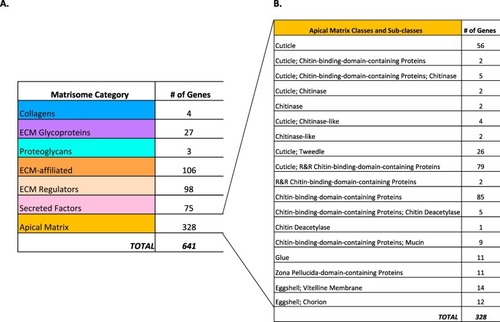
The Drosophila matrisome. (A) The Drosophila matrisome comprises 641 genes. These genes are then divided into either categories which we have previously defined, or the newly proposed apical matrix category. (B) The genes that encode proteins that make up the apical matrix of Drosophila were further divided into classes and sub-classes. |