- Title
-
Is zebrafish heart regeneration "complete"? Lineage-restricted cardiomyocytes proliferate to pre-injury numbers but some fail to differentiate in fibrotic hearts
- Authors
- Bertozzi, A., Wu, C.C., Nguyen, P.D., Vasudevarao, M.D., Mulaw, M.A., Koopman, C.D., de Boer, T.P., Bakkers, J., Weidinger, G.
- Source
- Full text @ Dev. Biol.
|
Fig. 1. Cardiomyocyte cell cycle activity drops to low levels prior to complete morphological regeneration. (A) The fraction of PCNA+ EGFP+ cardiomyocytes in myl7:EGFPtwu34 fish within 150 μm of the wound border peaks at 7 dpi and drops to levels barely above baseline by 30 dpi. uninj, uninjured. Error bars represent CI 95%. n = 5 hearts for each group. One-way ANOVA + Dunnett’s multiple comparisons test. Numbers indicate p-value. (B) Fraction of EdU+ EGFP+ cardiomyocytes in myl7:EGFPtwu34 fish. For each time point, a single intraperitoneal injection of EdU was performed 24 h prior to heart extraction. n (hearts) = 5 (uninjured), 5 (3 dpi), 5 (7 dpi), 7 (14 dpi), 8 (30 dpi), 7 (60 dpi). One-way ANOVA + Dunnett’s multiple comparisons test. (C) Many hearts contain wound tissue detected by Acid Fuchsin Orange G (AFOG) staining at 30 dpi. Red: fibrin, blue: collagen, brown: myocardium. Scale bars, 100 μm. (D) Percentage of hearts without residual wounds (= 0% of ventricle area), small wounds (between 0% and 3%) or bigger wounds (>3%) at 14, 30 and 90 dpi. n = 31 (14 dpi), 29 (30 dpi), 58 (90 dpi). Chi-square test. |
|
Fig. 2. Zebrafish hearts regenerate all cardiomyocytes lost to injury. (A) Experimental design for quantification of cardiomyocytes. (B) Representative images of −5.1myl7:DsRed2-nlsf2Tg hearts after sham injury or cryoinjury, at 3 or 90 days post surgery. Dashed yellow lines at 3 and 90 dpi outline the wound area. Scale bars, 100 μm. (C) Cardiomyocytes regenerate to the same number as found in sham injured hearts within 90 days. The total number of ventricular cardiomyocytes per heart is plotted. Error bars, CI 95%. n (hearts) = 99 (3 dps), 92 (90 dps), 68 (3 dpi), 58 (90 dpi). One-way ANOVA + Tukey’s multiple comparisons test. Observed difference 90 dpi vs 90 dps = 7400 (4% of CMs in 90 dps); smallest significant detectable difference = 19800 (12%). (D) Cardiomyocyte regeneration is not complete at 14 dpi. n (hearts) = 35 (3 dps), 35 (14 dps), 27 (3 dpi), 31 (14 dpi). One-way ANOVA + Tukey’s multiple comparisons test. (E) Pre-injury cardiomyocyte numbers have been restored at 30 dpi. n (hearts) = 24 (3 dps), 26 (30 dps), 27 (3 dpi), 29 (30 dpi). One-way ANOVA + Tukey’s multiple comparisons test. Observed difference 30 dpi vs 3 dps = 4100 (3.6% of CMs in 3 dps); smallest significant detectable difference = 17600 (15%). (F) The number of regenerated cardiomyocytes at 14, 30 and 90 dpi relative to the number missing in the respective 3 dpi group is plotted. Dotted line represents complete regeneration. n (hearts) = 31 (14 dpi), 29 (30 dpi), 58 (90 dpi). One-way ANOVA + Tukey’s multiple comparisons test. |
|
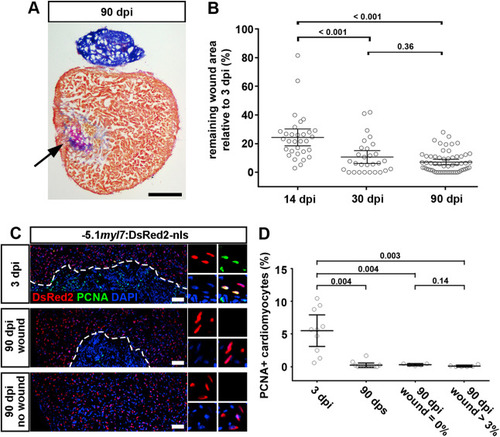
Fig. 3. Cardiomyocyte proliferation ceases in both scarred and unscarred hearts. (A) AFOG stained section at 90 dpi displaying incomplete scar resorption (arrow). Scale bar, 100 μm. (B) Average wound area does not significantly shrink between 30 and 90 dpi. n (hearts) = 31 (14 dpi), 29 (30 dpi), 58 (90 dpi). One-way ANOVA + Tukey’s multiple comparisons test. Observed difference 90 dpi vs 30 dpi = 3.5 (32% of 30 dpi wound area); smallest significant detectable difference = 5.8 (54% of 30 dpi wound area). (C, D) PCNA+ cardiomyocytes can be detected at 3 dpi, but not in 90 dpi hearts with or without wound. Scale bar, 100 μm n (hearts) = 10 (3 dpi), 12 (90 dps), 5 (90 dpi no wound), 6 (90 dpi wound). One-way ANOVA + Tukey’s multiple comparisons test. |
|
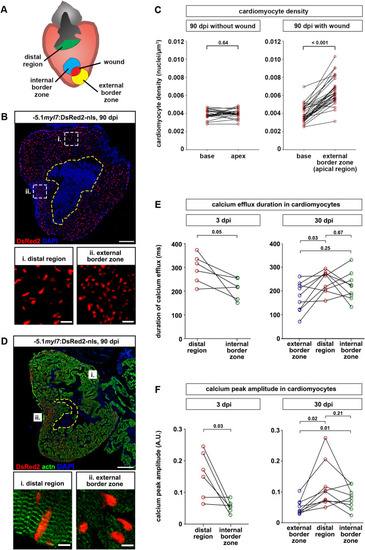
Fig. 4. Cardiomyocytes are denser and less mature at the external wound border zone in scarred hearts. (A) Cartoon depicting different regions of ventricles containing residual scars that were analyzed. (B) Cardiomyocyte nuclei density is higher in the external wound border zone (box ii) than the distal region (box i) at 90 dpi. The wound is outlined with a dashed yellow line. Scale bar, 100 μm. (C) At 90 dpi, cardiomyocyte density at the base and apex does not differ in hearts without wound, but is higher at the external wound border in hearts with wounds. n (hearts) = 22 (without wound), 30 (wound). Paired two-tailed t-test. (D) Sarcomeric structures cannot be identified by alpha-actinin staining in the external wound border in hearts with residual scars. n (hearts with assembled sarcomeres): wound border, 2/13 hearts at 30 dpi, 4/23 hearts at 90 dpi; distal myocardium, 12/13 hearts at 30 dpi, 21/23 hearts at 90 dpi. The wound is outlined with a dashed yellow line. Scale bar, 100 μm. (E, F) Calcium efflux duration (E) and amplitude of calcium peaks (F) in myl7:GCaMP6f-nls-T2A-RCaMP107-NES-pA)hu11799 cardiomyocytes is plotted at 3 dpi and at 30 dpi for hearts containing residual scars. n (hearts) = 6 (3 dpi), 9 (30 dpi). Paired two-tailed t-test. |
|
Fig. 5. Cardiomyocytes remain lineage-restricted during adult heart regeneration. (A) Schematic of the hsp70l:LOXP-luc-myc-STOP-LOXP-dTomato, cryaa:YPetulm9 (hsp:L to T) transgene. (B–H) At 7 dpi, myc-tagged luciferase is not detectable in the ventricle of adult hsp70l:LOXP-luc-myc-STOP-LOXP-dTomato, cryaa:YPetulm9 (hsp:L to T) hearts without heat-shocks (B). After heat-shock, expression is found in myl7+ cardiomyocytes (C), Et(krt4:eGFPsqet27)+ epicardial cells (D), fli1a:eGFPy1Tg+endocardial cells (E), coro1a:eGFPhkz04tTg+leukocytes (F), in wt1b:EGFPli1Tg+epicardial cells, fibroblasts and macrophages (G), and in postnb:Citrinecn6Tg+ activated fibroblasts (H). Boxed areas are shown in magnified view. Scale bars, 100 μm (overview), 10 μm (magnified view). (I) Experimental design for adult cardiomyocyte lineage tracing. (J) dTomato+ traced cells are randomly located throughout the ventricle, and express the cardiomyocyte marker myosin heavy chain (MHC). Scale bars, 100 μm (overview), 12.5 μm (magnified view). (K) No difference is detected in average percentages of dTomato+ traced cells expressing MHC between uninjured and regenerating hearts. Error bars, s.e.m. n (hearts) = 4 (uninjured), 4 (7 dpi), 5 (30 dpi). n (traced cells) = 942 (uninjured), 775 (7 dpi), 1134 (30 dpi). One-way ANOVA + Holm-Sidak’s multiple comparisons test. Observed difference uninj. vs 7 dpi = 0.3 (0.3% of traced cells in uninj.); smallest significant detectable difference = 0.7 (0.7% of uninj.). Observed difference uninj. vs 30 dpi = 0.6 (0.6% of traced cells in uninj.); smallest significant detectable difference = 1.2 (1.2% of uninj.). Observed difference 7 dpi vs 30 dpi = 0.2 (0.2% of traced cells in 7 dpi); smallest significant detectable difference = 1.3 (1.4% of 7 dpi). |
|
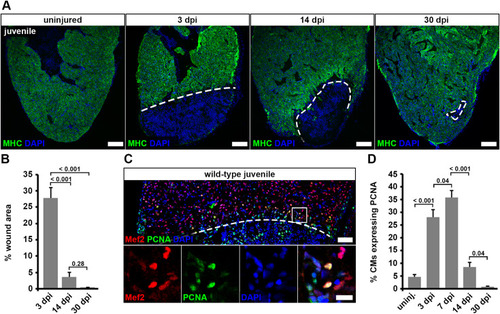
Fig. 6. Juvenile hearts regenerate after cryoinjury via cardiomyocyte proliferation. (A) Myosin heavy chain (MHC) immunostaining reveals wound area (dashed lines) on heart sections of cryoinjured juvenile fish. Scale bars, 100 μm. (B) Wound area (MHC— area) relative to the whole ventricle area is plotted. Error bars represent s.e.m. n (hearts) = 6 for all time-points. One-way ANOVA + Holm-Sidak’s multiple comparisons test. (C) PCNA+ Mef2+ cardiomyocytes in juvenile hearts at 7 dpi. Scale bars, 50 μm (overview), 12.5 μm (magnified view). (D) The average percentage of cardiomyocytes expressing PCNA within 150 μm from the wound border is plotted. Error bars represent s.e.m. n (hearts) = 6 for all time-points. One-way ANOVA + Holm-Sidak’s multiple comparisons test. |
|
Fig. 7. Cardiomyocytes remain lineage-restricted during juvenile heart regeneration. (A–D) At 7 dpi in juvenile fish, expression of the hsp:R to nGtud9 tracer transgene is detected by DsRed immunofluorescence after heat-shock (h.s.) in endocardial cells (Aldh1a2+, boxed area i in (A)), epicardial cells (Aldh1a2+, boxed area ii in (A)), blood cells (Aldh1a2— with small, round and intense DAPI-stained nuclei, boxed area iii in (A)), cardiomyocytes (Myl7+, boxed area in (B)), wt1b:EGFPli1Tg+epicardial cells, fibroblasts and macrophages (C), and in postnb:Citrinecn6Tg+fibroblasts (D). Scale bars, 40 μm (overview), 5 μm (magnified view). (E) Experimental design for juvenile cardiomyocyte lineage tracing. (F) Nuclear GFP+ traced cells are widespread throughout the ventricle and colocalize with Mef2, a nuclear cardiomyocyte marker. Scale bars, 100 μm (overview), 12.5 μm (magnified view). (G) No significant difference is detected in average percentages of GFP+ cells expressing Mef2 between uninjured and regenerating hearts. Error bars, s.e.m. n (hearts) = 4 (uninjured), 5 (7 dpi), 6 (30 dpi). n (traced cells) = 858 (uninjured), 823 (7 dpi), 1123 (30 dpi). One-way ANOVA + Holm-Sidak’s multiple comparisons test. Observed difference uninj. vs 7 dpi = 0.06 (0.06% of traced cells in uninj.); smallest significant detectable difference = 0.8 (0.8% of uninj.). Observed difference uninj. vs 30 dpi = 0.8 (0.8% of traced cells in uninj.); smallest significant detectable difference = 1.9 (1.9% of uninj.). Observed difference 7 dpi vs 30 dpi = 0.9 (0.9% of traced cells in 7 dpi); smallest significant detectable difference = 1.6 (1.6% of 7 dpi). |
|
Fig. 8. Quantitative model of zebrafish heart regeneration via proliferation of lineage-restricted cardiomyocytes. (A) Model of cardiomyocyte fate during zebrafish heart regeneration. Cardiomyocytes dedifferentiate, which entails loss of sarcomeres, alterations in calcium transients and gain of embryonic markers, but not attainment of multipotency. (B) Model of the time-courses of cardiomyocyte regeneration and wound resorption. Restoration of pre-injury cardiomyocyte numbers (green) in response to cryoinjury, which kills about 30% of the cardiomyocytes, is achieved within 30 days. The morphological septation between trabecular layer (pink) and cortical layer (brown) is restored. Despite completed cardiomyocyte regeneration, the majority of hearts still contain scars at 30 and 90 dpi. Regenerated cardiomyocytes in scar-free ventricles or at the internal wound border zone retain normal density and fully re-differentiate (blue). Some of the regenerated cardiomyocytes (3–15%) located externally to remaining internalized scars are smaller and only partially re-differentiated (yellow). In scarred hearts, fibrosis persists beyond 30 dpi. |
Reprinted from Developmental Biology, 471, Bertozzi, A., Wu, C.C., Nguyen, P.D., Vasudevarao, M.D., Mulaw, M.A., Koopman, C.D., de Boer, T.P., Bakkers, J., Weidinger, G., Is zebrafish heart regeneration "complete"? Lineage-restricted cardiomyocytes proliferate to pre-injury numbers but some fail to differentiate in fibrotic hearts, 106-118, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.