- Title
-
Colored visual stimuli evoke spectrally tuned neuronal responses across the central nervous system of zebrafish larvae
- Authors
- Fornetto, C., Tiso, N., Pavone, F.S., Vanzi, F.
- Source
- Full text @ BMC Biol.
|
Experimental setup, visual stimulation protocol, and regression-based identification of responsive neurons. |
|
|
|
Stimulus-induced neuronal activity: spectral and anatomical mapping. |
|
Spectrally tuned responses and anatomical distributions. Δ |
|
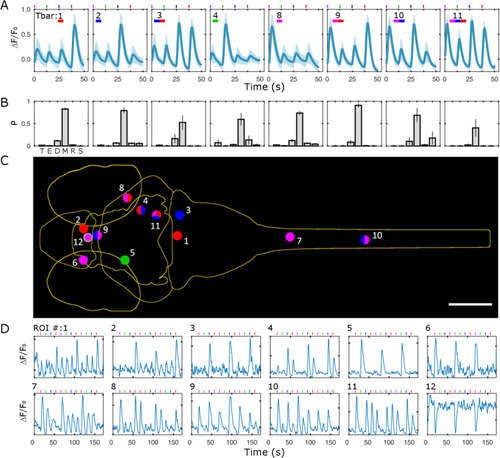
Unique spectral classification with Tbar. |
|
CNS spectral and anatomical mapping in 3-dpf larvae. |