- Title
-
Human ARF Specifically Inhibits Epimorphic Regeneration in the Zebrafish Heart
- Authors
- Lee, S., Hesse, R., Tamaki, S., Garland, C., Pomerantz, J.H.
- Source
- Full text @ Genes (Basel)
|
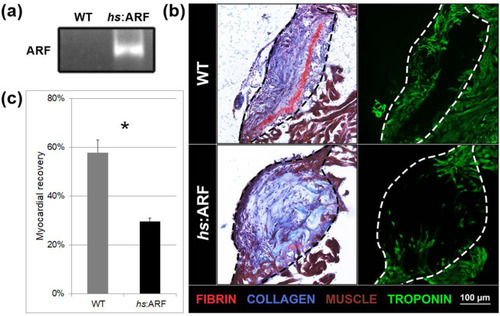
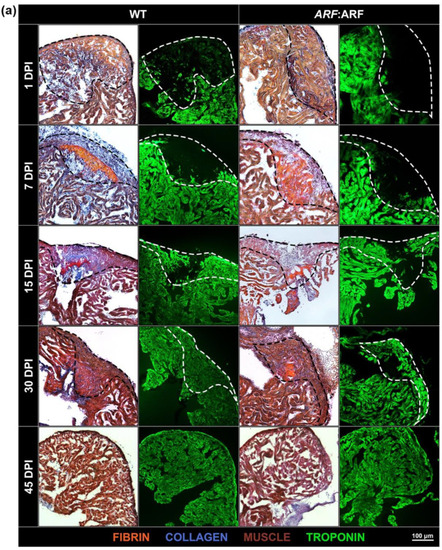
Induced Alternative Reading Frame (ARF) expression in |
|
ARF expressed under control of the endogenous human |
|
ARF expressed under control of the endogenous human |
|
ARF expressed in PHENOTYPE:
|
|
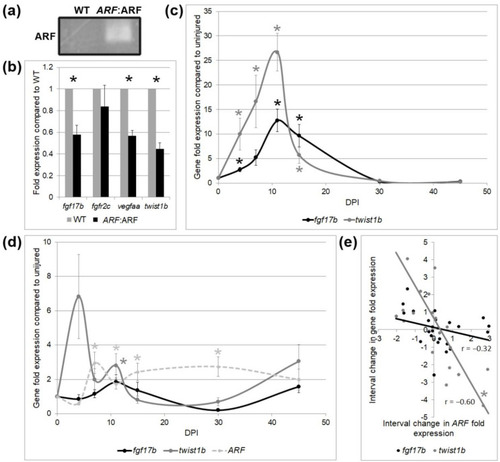
Tissue-specific gene expression by qPCR in WT and |